Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
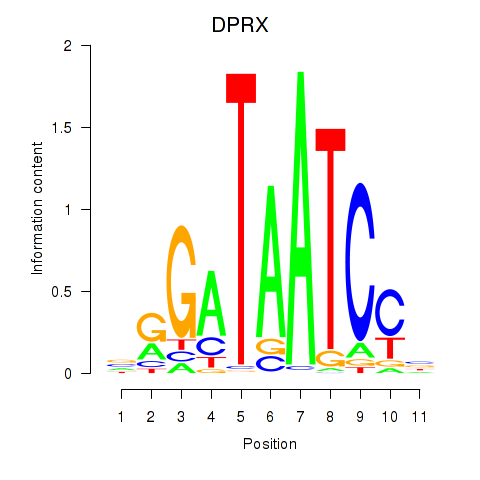
Results for DPRX
Z-value: 0.51
Transcription factors associated with DPRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DPRX
|
ENSG00000204595.1 | divergent-paired related homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DPRX | hg19_v2_chr19_+_54135310_54135310 | 0.89 | 1.9e-02 | Click! |
Activity profile of DPRX motif
Sorted Z-values of DPRX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_321340 | 1.04 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr11_+_61248583 | 0.32 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr11_-_321050 | 0.31 |
ENST00000399808.4
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr12_+_113344582 | 0.29 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr16_+_30386098 | 0.25 |
ENST00000322861.7
|
MYLPF
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr9_+_116343192 | 0.24 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr1_-_149400542 | 0.23 |
ENST00000392948.2
|
HIST2H3PS2
|
histone cluster 2, H3, pseudogene 2 |
| chr17_+_77021702 | 0.21 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr19_+_21106081 | 0.21 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr5_+_136070614 | 0.21 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr2_-_68290106 | 0.20 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chrX_+_115567767 | 0.19 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr11_-_61596753 | 0.19 |
ENST00000448607.1
ENST00000421879.1 |
FADS1
|
fatty acid desaturase 1 |
| chr2_-_26251481 | 0.19 |
ENST00000599234.1
|
AC013449.1
|
Uncharacterized protein |
| chr1_+_169079823 | 0.19 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr15_+_96904487 | 0.17 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr8_-_42360015 | 0.17 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_-_150460914 | 0.16 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_+_100435535 | 0.16 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr6_-_26108355 | 0.16 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr12_-_49523896 | 0.15 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr18_-_64271316 | 0.15 |
ENST00000540086.1
ENST00000580157.1 |
CDH19
|
cadherin 19, type 2 |
| chr8_-_93978309 | 0.14 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr20_-_36794902 | 0.13 |
ENST00000373403.3
|
TGM2
|
transglutaminase 2 |
| chr15_-_42565221 | 0.13 |
ENST00000563371.1
ENST00000568400.1 ENST00000568432.1 |
TMEM87A
|
transmembrane protein 87A |
| chr8_-_71520513 | 0.13 |
ENST00000262213.2
ENST00000536748.1 ENST00000518678.1 |
TRAM1
|
translocation associated membrane protein 1 |
| chr12_+_113344811 | 0.12 |
ENST00000551241.1
ENST00000553185.1 ENST00000550689.1 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_+_141105705 | 0.12 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr5_+_35856951 | 0.12 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr16_-_74734742 | 0.12 |
ENST00000308807.7
ENST00000573267.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr5_+_150040403 | 0.11 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr19_+_41284121 | 0.11 |
ENST00000594800.1
ENST00000357052.2 ENST00000602173.1 |
RAB4B
|
RAB4B, member RAS oncogene family |
| chr3_+_111393501 | 0.11 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr7_+_89975979 | 0.11 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr3_-_32612263 | 0.11 |
ENST00000432458.2
ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr20_-_36794938 | 0.11 |
ENST00000453095.1
|
TGM2
|
transglutaminase 2 |
| chr1_+_200863949 | 0.11 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr1_-_46089639 | 0.11 |
ENST00000445048.2
|
CCDC17
|
coiled-coil domain containing 17 |
| chr19_-_57967854 | 0.10 |
ENST00000321039.3
|
VN1R1
|
vomeronasal 1 receptor 1 |
| chr7_+_28452130 | 0.10 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr12_+_93772402 | 0.10 |
ENST00000546925.1
|
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr11_-_61849656 | 0.10 |
ENST00000524942.1
|
RP11-810P12.5
|
RP11-810P12.5 |
| chr8_+_101349823 | 0.10 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr7_-_135412925 | 0.10 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr8_+_94752349 | 0.10 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr12_-_124419531 | 0.10 |
ENST00000514254.2
|
DNAH10OS
|
dynein, axonemal, heavy chain 10 opposite strand |
| chr8_-_95274536 | 0.10 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr8_-_93978333 | 0.09 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr4_+_84377198 | 0.09 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr16_-_71323271 | 0.09 |
ENST00000565850.1
ENST00000568910.1 ENST00000434935.2 ENST00000338099.5 |
CMTR2
|
cap methyltransferase 2 |
| chr7_+_7222157 | 0.09 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr15_-_44092295 | 0.09 |
ENST00000249714.3
ENST00000299969.6 ENST00000319327.6 |
SERINC4
|
serine incorporator 4 |
| chr1_-_173174681 | 0.09 |
ENST00000367718.1
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr12_+_93772326 | 0.09 |
ENST00000550056.1
ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr16_-_46649905 | 0.09 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr17_-_4643114 | 0.09 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr7_-_75401513 | 0.09 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr4_-_87279520 | 0.08 |
ENST00000506773.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr11_-_65629497 | 0.08 |
ENST00000532134.1
|
CFL1
|
cofilin 1 (non-muscle) |
| chr11_-_8816375 | 0.08 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr6_-_32908765 | 0.08 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr5_+_147691979 | 0.08 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr4_+_89299885 | 0.08 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr5_+_118604385 | 0.08 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_+_232063436 | 0.08 |
ENST00000440107.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr17_-_47925379 | 0.08 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr12_+_104359576 | 0.08 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr12_+_6949964 | 0.07 |
ENST00000541978.1
ENST00000435982.2 |
GNB3
|
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr3_+_40351169 | 0.07 |
ENST00000232905.3
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr8_-_37411648 | 0.07 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr3_-_178976996 | 0.07 |
ENST00000485523.1
|
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr5_+_94955782 | 0.07 |
ENST00000380007.2
|
GPR150
|
G protein-coupled receptor 150 |
| chr1_+_36621174 | 0.07 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr16_-_74734672 | 0.07 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr18_+_32290218 | 0.07 |
ENST00000348997.5
ENST00000588949.1 ENST00000597599.1 |
DTNA
|
dystrobrevin, alpha |
| chr19_+_48325097 | 0.07 |
ENST00000221996.7
ENST00000539067.1 |
CRX
|
cone-rod homeobox |
| chr12_-_111395610 | 0.07 |
ENST00000548329.1
ENST00000546852.1 |
RP1-46F2.3
|
RP1-46F2.3 |
| chr4_-_87279641 | 0.07 |
ENST00000512689.1
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_113574028 | 0.07 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr1_-_150780757 | 0.07 |
ENST00000271651.3
|
CTSK
|
cathepsin K |
| chr8_-_93978346 | 0.07 |
ENST00000523580.1
|
TRIQK
|
triple QxxK/R motif containing |
| chr17_+_7531281 | 0.07 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr8_-_80993010 | 0.07 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr19_-_44123734 | 0.07 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr19_-_44124019 | 0.07 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr4_-_87374330 | 0.07 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_89300158 | 0.06 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_100435315 | 0.06 |
ENST00000370155.3
ENST00000465289.1 |
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr8_-_124037890 | 0.06 |
ENST00000519018.1
ENST00000523036.1 |
DERL1
|
derlin 1 |
| chr16_-_28415162 | 0.06 |
ENST00000398944.3
ENST00000398943.3 ENST00000380876.4 |
EIF3CL
|
eukaryotic translation initiation factor 3, subunit C-like |
| chr7_+_7222233 | 0.06 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr16_+_28722684 | 0.06 |
ENST00000331666.6
ENST00000395587.1 ENST00000569690.1 ENST00000564243.1 |
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chrX_-_49121165 | 0.06 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chrX_+_15767971 | 0.06 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr4_+_108911036 | 0.06 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr19_+_36157715 | 0.06 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr6_-_33290580 | 0.06 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr12_+_104359614 | 0.06 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr16_+_28722809 | 0.06 |
ENST00000566866.1
|
EIF3C
|
eukaryotic translation initiation factor 3, subunit C |
| chr10_+_118187379 | 0.06 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr12_-_96184913 | 0.06 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr16_-_18911366 | 0.06 |
ENST00000565224.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr12_+_113416265 | 0.06 |
ENST00000449768.2
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr1_+_43148059 | 0.06 |
ENST00000321358.7
ENST00000332220.6 |
YBX1
|
Y box binding protein 1 |
| chr5_+_63461642 | 0.06 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr15_+_29211570 | 0.06 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr4_-_47465666 | 0.05 |
ENST00000381571.4
|
COMMD8
|
COMM domain containing 8 |
| chr14_+_61995722 | 0.05 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr8_+_72755367 | 0.05 |
ENST00000537896.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr3_+_188664988 | 0.05 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr7_-_97881429 | 0.05 |
ENST00000420697.1
ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1
|
tectonin beta-propeller repeat containing 1 |
| chr14_+_59951161 | 0.05 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chrX_+_100878079 | 0.05 |
ENST00000471229.2
|
ARMCX3
|
armadillo repeat containing, X-linked 3 |
| chr10_-_112255945 | 0.05 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr15_+_63481668 | 0.05 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr19_-_12267524 | 0.05 |
ENST00000455799.1
ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625
|
zinc finger protein 625 |
| chr8_-_93978357 | 0.05 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chrX_-_15683147 | 0.05 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr1_-_55089191 | 0.05 |
ENST00000302250.2
ENST00000371304.2 |
FAM151A
|
family with sequence similarity 151, member A |
| chr1_-_762885 | 0.05 |
ENST00000536430.1
ENST00000473798.1 |
LINC00115
|
long intergenic non-protein coding RNA 115 |
| chr1_+_74701062 | 0.05 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr1_-_115301235 | 0.05 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr20_+_361890 | 0.05 |
ENST00000449710.1
ENST00000422053.2 |
TRIB3
|
tribbles pseudokinase 3 |
| chr1_+_16767167 | 0.05 |
ENST00000337132.5
|
NECAP2
|
NECAP endocytosis associated 2 |
| chr1_+_207262881 | 0.05 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr3_-_135913298 | 0.05 |
ENST00000434835.2
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr8_-_79717750 | 0.04 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr11_+_108094786 | 0.04 |
ENST00000601453.1
|
ATM
|
ataxia telangiectasia mutated |
| chrX_+_118425471 | 0.04 |
ENST00000428222.1
|
RP5-1139I1.1
|
RP5-1139I1.1 |
| chr17_-_4448361 | 0.04 |
ENST00000572759.1
|
MYBBP1A
|
MYB binding protein (P160) 1a |
| chr4_+_108910870 | 0.04 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr11_+_107879459 | 0.04 |
ENST00000393094.2
|
CUL5
|
cullin 5 |
| chr11_-_10715287 | 0.04 |
ENST00000423302.2
|
MRVI1
|
murine retrovirus integration site 1 homolog |
| chr5_+_78365577 | 0.04 |
ENST00000518666.1
ENST00000521567.1 |
BHMT2
|
betaine--homocysteine S-methyltransferase 2 |
| chr1_+_36772691 | 0.04 |
ENST00000312808.4
ENST00000505871.1 |
SH3D21
|
SH3 domain containing 21 |
| chr8_-_53626974 | 0.04 |
ENST00000435644.2
ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1
|
RB1-inducible coiled-coil 1 |
| chr1_+_36024107 | 0.04 |
ENST00000437806.1
|
NCDN
|
neurochondrin |
| chr5_-_77072085 | 0.04 |
ENST00000518338.2
ENST00000520039.1 ENST00000306388.6 ENST00000520361.1 |
TBCA
|
tubulin folding cofactor A |
| chr4_-_175205407 | 0.04 |
ENST00000393674.2
|
FBXO8
|
F-box protein 8 |
| chr19_-_18709357 | 0.04 |
ENST00000597131.1
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr13_-_45775162 | 0.04 |
ENST00000405872.1
|
KCTD4
|
potassium channel tetramerization domain containing 4 |
| chr12_+_86268065 | 0.04 |
ENST00000551529.1
ENST00000256010.6 |
NTS
|
neurotensin |
| chr1_-_94586651 | 0.04 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr16_+_30406423 | 0.04 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr5_-_77072145 | 0.04 |
ENST00000380377.4
|
TBCA
|
tubulin folding cofactor A |
| chr12_-_56105880 | 0.04 |
ENST00000557257.1
|
ITGA7
|
integrin, alpha 7 |
| chr5_-_117897751 | 0.04 |
ENST00000515704.1
ENST00000506769.1 |
CTD-2281M20.1
|
CTD-2281M20.1 |
| chr3_+_97483366 | 0.04 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr11_-_64949305 | 0.04 |
ENST00000526623.1
|
AP003068.23
|
Uncharacterized protein |
| chr11_+_63449045 | 0.04 |
ENST00000354497.4
|
RTN3
|
reticulon 3 |
| chr9_+_17135016 | 0.04 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr17_-_73150629 | 0.04 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr17_-_5323480 | 0.04 |
ENST00000573584.1
|
NUP88
|
nucleoporin 88kDa |
| chr1_-_150208320 | 0.04 |
ENST00000534220.1
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr4_+_153701081 | 0.04 |
ENST00000451320.2
ENST00000429148.2 ENST00000353617.2 ENST00000405727.2 ENST00000356064.3 |
ARFIP1
|
ADP-ribosylation factor interacting protein 1 |
| chr16_+_57844549 | 0.03 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr19_+_49999631 | 0.03 |
ENST00000270625.2
ENST00000596873.1 ENST00000594493.1 ENST00000599561.1 |
RPS11
|
ribosomal protein S11 |
| chr16_-_21416640 | 0.03 |
ENST00000542817.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr14_-_101034811 | 0.03 |
ENST00000553553.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr8_+_145215928 | 0.03 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr2_+_114737146 | 0.03 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chrX_+_153769409 | 0.03 |
ENST00000440286.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr17_+_8191815 | 0.03 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr4_-_164534657 | 0.03 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_-_45988542 | 0.03 |
ENST00000424390.1
|
PRDX1
|
peroxiredoxin 1 |
| chr11_+_120973375 | 0.03 |
ENST00000264037.2
|
TECTA
|
tectorin alpha |
| chr16_-_15950868 | 0.03 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr18_-_67614645 | 0.03 |
ENST00000577287.1
|
CD226
|
CD226 molecule |
| chr9_-_132404374 | 0.03 |
ENST00000277459.4
ENST00000450050.2 ENST00000277458.4 |
ASB6
|
ankyrin repeat and SOCS box containing 6 |
| chr5_-_59064458 | 0.03 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_-_93978216 | 0.03 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr1_+_182808474 | 0.03 |
ENST00000367549.3
|
DHX9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr4_+_69313145 | 0.03 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr17_-_42019836 | 0.03 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr1_+_16767195 | 0.03 |
ENST00000504551.2
ENST00000457722.2 ENST00000406746.1 ENST00000443980.2 |
NECAP2
|
NECAP endocytosis associated 2 |
| chr1_+_36023370 | 0.03 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr8_+_38244638 | 0.03 |
ENST00000526356.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr6_+_55192267 | 0.03 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr1_+_19638788 | 0.03 |
ENST00000375155.3
ENST00000375153.3 ENST00000400548.2 |
PQLC2
|
PQ loop repeat containing 2 |
| chr14_-_101034407 | 0.03 |
ENST00000443071.2
ENST00000557378.1 |
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr16_-_30022735 | 0.03 |
ENST00000564944.1
|
DOC2A
|
double C2-like domains, alpha |
| chr12_+_104359641 | 0.03 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr10_+_51572408 | 0.03 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr5_-_159846066 | 0.03 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr2_+_234216454 | 0.03 |
ENST00000447536.1
ENST00000409110.1 |
SAG
|
S-antigen; retina and pineal gland (arrestin) |
| chr17_-_48785216 | 0.03 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr21_-_28217721 | 0.03 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr12_+_56552128 | 0.03 |
ENST00000548580.1
ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr3_+_111393659 | 0.02 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr11_+_65343494 | 0.02 |
ENST00000309295.4
ENST00000533237.1 |
EHBP1L1
|
EH domain binding protein 1-like 1 |
| chr15_-_42565606 | 0.02 |
ENST00000307216.6
ENST00000448392.1 |
TMEM87A
|
transmembrane protein 87A |
| chr6_+_26383404 | 0.02 |
ENST00000416795.2
ENST00000494184.1 |
BTN2A2
|
butyrophilin, subfamily 2, member A2 |
| chr1_-_110950255 | 0.02 |
ENST00000483260.1
ENST00000474861.2 ENST00000602318.1 |
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr17_+_48351785 | 0.02 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr2_+_113342163 | 0.02 |
ENST00000409719.1
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_+_5323492 | 0.02 |
ENST00000405578.4
ENST00000574003.1 |
RPAIN
|
RPA interacting protein |
| chr20_+_22034809 | 0.02 |
ENST00000449427.1
|
RP11-125P18.1
|
RP11-125P18.1 |
| chr6_-_37225367 | 0.02 |
ENST00000336655.2
|
TMEM217
|
transmembrane protein 217 |
| chr2_-_176046391 | 0.02 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr12_+_130554803 | 0.02 |
ENST00000535487.1
|
RP11-474D1.2
|
RP11-474D1.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DPRX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 1.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.2 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:1903281 | positive regulation of calcium:sodium antiporter activity(GO:1903281) |
| 0.0 | 0.2 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:2000769 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.0 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.1 | 0.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.2 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.0 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |