Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
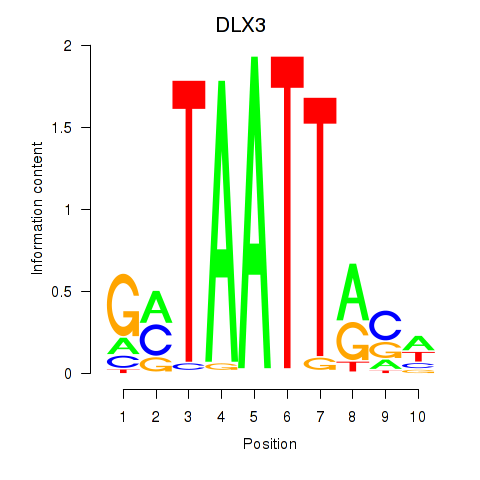
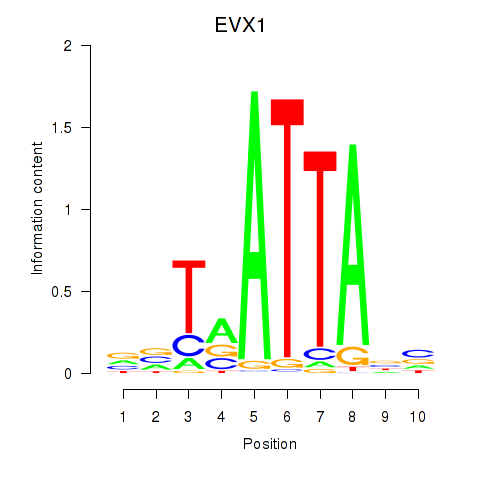
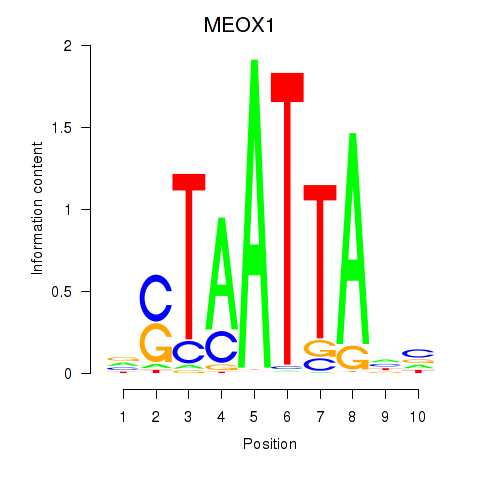
Results for DLX3_EVX1_MEOX1
Z-value: 0.34
Transcription factors associated with DLX3_EVX1_MEOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX3
|
ENSG00000064195.7 | distal-less homeobox 3 |
|
EVX1
|
ENSG00000106038.8 | even-skipped homeobox 1 |
|
MEOX1
|
ENSG00000005102.8 | mesenchyme homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEOX1 | hg19_v2_chr17_-_41738931_41739040 | 0.53 | 2.8e-01 | Click! |
| DLX3 | hg19_v2_chr17_-_48072574_48072597 | -0.32 | 5.3e-01 | Click! |
Activity profile of DLX3_EVX1_MEOX1 motif
Sorted Z-values of DLX3_EVX1_MEOX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_57229155 | 0.38 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr13_-_30683005 | 0.29 |
ENST00000413591.1
ENST00000432770.1 |
LINC00365
|
long intergenic non-protein coding RNA 365 |
| chr2_+_17997763 | 0.27 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr12_-_89746264 | 0.24 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr16_+_53133070 | 0.23 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr3_-_143567262 | 0.23 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr7_-_99716914 | 0.19 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_99716952 | 0.19 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_+_37723336 | 0.16 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr5_+_66300464 | 0.16 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr6_-_116833500 | 0.15 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr12_+_78359999 | 0.13 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr15_+_75080883 | 0.13 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr7_-_99717463 | 0.13 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr7_-_99716940 | 0.12 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_-_203735484 | 0.11 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chrX_-_100129128 | 0.11 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr5_+_66300446 | 0.11 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_42234745 | 0.11 |
ENST00000220812.2
|
DKK4
|
dickkopf WNT signaling pathway inhibitor 4 |
| chr12_+_28410128 | 0.10 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr14_-_37051798 | 0.10 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr16_+_53412368 | 0.10 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr7_-_33102338 | 0.09 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr2_+_196313239 | 0.09 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr1_-_21625486 | 0.09 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr5_+_174151536 | 0.09 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr7_+_80253387 | 0.09 |
ENST00000438020.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr1_-_159832438 | 0.08 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr17_-_71223839 | 0.08 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr6_-_75994025 | 0.08 |
ENST00000518161.1
|
TMEM30A
|
transmembrane protein 30A |
| chr11_-_31531121 | 0.08 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr17_-_48133054 | 0.08 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr17_+_62223320 | 0.08 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr11_-_26593779 | 0.07 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr7_-_111424506 | 0.07 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr13_+_78315528 | 0.07 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr10_-_21186144 | 0.07 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_-_94673956 | 0.06 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr7_+_150811705 | 0.06 |
ENST00000335367.3
|
AGAP3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr9_-_5339873 | 0.06 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr15_+_59910132 | 0.06 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr14_+_97925151 | 0.06 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr4_-_105416039 | 0.06 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr7_-_33102399 | 0.06 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr4_-_48683188 | 0.06 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr18_-_33709268 | 0.06 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr1_-_234667504 | 0.06 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr17_-_33446735 | 0.06 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr19_-_7968427 | 0.06 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chrM_+_8366 | 0.06 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr5_-_115890554 | 0.05 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr17_+_73539339 | 0.05 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr12_-_719573 | 0.05 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr3_+_2933893 | 0.05 |
ENST00000397459.2
|
CNTN4
|
contactin 4 |
| chr7_-_100860851 | 0.05 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_-_50889155 | 0.05 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr17_-_77924627 | 0.05 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr20_+_60174827 | 0.05 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr18_-_68004529 | 0.05 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr1_+_28586006 | 0.04 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr9_-_136933615 | 0.04 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr5_-_87516448 | 0.04 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr20_-_61733657 | 0.04 |
ENST00000608031.1
ENST00000447910.2 |
HAR1B
|
highly accelerated region 1B (non-protein coding) |
| chr15_+_63188009 | 0.04 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr10_+_18549645 | 0.04 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr5_-_141249154 | 0.04 |
ENST00000357517.5
ENST00000536585.1 |
PCDH1
|
protocadherin 1 |
| chr13_+_110958124 | 0.04 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr1_+_36621174 | 0.04 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr4_-_36245561 | 0.04 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr18_-_56985776 | 0.04 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr5_-_141030943 | 0.04 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr16_+_30675654 | 0.04 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr13_-_38172863 | 0.04 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr14_-_90085458 | 0.04 |
ENST00000345097.4
ENST00000555855.1 ENST00000555353.1 |
FOXN3
|
forkhead box N3 |
| chr20_+_43990576 | 0.04 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr7_+_80255472 | 0.04 |
ENST00000428497.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr15_+_89164560 | 0.04 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr8_+_22424551 | 0.04 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr4_+_105828537 | 0.04 |
ENST00000515649.1
|
RP11-556I14.1
|
RP11-556I14.1 |
| chr19_-_4902877 | 0.03 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr7_+_73245193 | 0.03 |
ENST00000340958.2
|
CLDN4
|
claudin 4 |
| chr5_-_34043310 | 0.03 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr15_-_64673630 | 0.03 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr2_-_37544209 | 0.03 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr6_+_43968306 | 0.03 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chrX_-_23926004 | 0.03 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr17_+_42264556 | 0.03 |
ENST00000319511.6
ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr15_-_68497657 | 0.03 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chrX_+_133930798 | 0.03 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr16_+_57139933 | 0.03 |
ENST00000566259.1
|
CPNE2
|
copine II |
| chr22_-_41682172 | 0.03 |
ENST00000356244.3
|
RANGAP1
|
Ran GTPase activating protein 1 |
| chr4_+_77356248 | 0.03 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr1_-_197115818 | 0.03 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr7_+_75931861 | 0.03 |
ENST00000248553.6
|
HSPB1
|
heat shock 27kDa protein 1 |
| chr18_-_53019208 | 0.03 |
ENST00000562607.1
|
TCF4
|
transcription factor 4 |
| chr11_-_26593677 | 0.03 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chr19_+_34287751 | 0.03 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr13_-_34250861 | 0.03 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr9_-_136933134 | 0.03 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr2_+_149974684 | 0.03 |
ENST00000450639.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr17_-_39165366 | 0.03 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr2_+_113763031 | 0.03 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr9_+_130026756 | 0.03 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr16_+_14280564 | 0.03 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr3_-_108248169 | 0.03 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr3_-_195538760 | 0.03 |
ENST00000475231.1
|
MUC4
|
mucin 4, cell surface associated |
| chr2_+_201754050 | 0.03 |
ENST00000426253.1
ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr4_-_109541539 | 0.03 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr10_+_118187379 | 0.03 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr2_-_17981462 | 0.03 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr13_-_46716969 | 0.02 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr19_-_45681482 | 0.02 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr20_+_3776371 | 0.02 |
ENST00000245960.5
|
CDC25B
|
cell division cycle 25B |
| chr14_+_38264418 | 0.02 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr10_-_99447024 | 0.02 |
ENST00000370626.3
|
AVPI1
|
arginine vasopressin-induced 1 |
| chr11_-_70672645 | 0.02 |
ENST00000423696.2
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_-_39979576 | 0.02 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr3_+_44840679 | 0.02 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr16_-_29910853 | 0.02 |
ENST00000308713.5
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2 |
| chr11_+_128563652 | 0.02 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_34204642 | 0.02 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_-_73460334 | 0.02 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr7_-_151217166 | 0.02 |
ENST00000496004.1
|
RHEB
|
Ras homolog enriched in brain |
| chr2_-_61697862 | 0.02 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr14_+_64680854 | 0.02 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chrX_-_73051037 | 0.02 |
ENST00000445814.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr6_+_46761118 | 0.02 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr9_-_5304432 | 0.02 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr6_+_126221034 | 0.02 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr3_+_115342349 | 0.02 |
ENST00000393780.3
|
GAP43
|
growth associated protein 43 |
| chr3_-_47934234 | 0.02 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr6_+_160211481 | 0.02 |
ENST00000367034.4
|
MRPL18
|
mitochondrial ribosomal protein L18 |
| chr2_-_201753859 | 0.02 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr19_+_50016610 | 0.02 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr2_+_201754135 | 0.02 |
ENST00000409357.1
ENST00000409129.2 |
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr21_-_35899113 | 0.02 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr8_-_42358742 | 0.02 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr11_-_111794446 | 0.02 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr9_+_124329336 | 0.02 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chrX_+_10031499 | 0.02 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr16_-_69390209 | 0.02 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr3_-_33686743 | 0.02 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_-_47546220 | 0.02 |
ENST00000528538.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr11_-_117747327 | 0.02 |
ENST00000584230.1
ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6
FXYD6-FXYD2
|
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr4_-_174451370 | 0.02 |
ENST00000359562.4
|
HAND2
|
heart and neural crest derivatives expressed 2 |
| chr15_-_55563072 | 0.02 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_+_11071546 | 0.02 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr5_+_159343688 | 0.02 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr17_-_72772425 | 0.02 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_+_29001711 | 0.02 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr3_-_33686925 | 0.02 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_-_87028478 | 0.02 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr12_-_6233828 | 0.02 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr18_+_55888767 | 0.02 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr11_+_101918153 | 0.02 |
ENST00000434758.2
ENST00000526781.1 ENST00000534360.1 |
C11orf70
|
chromosome 11 open reading frame 70 |
| chr4_-_120243545 | 0.02 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chr14_-_36988882 | 0.02 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr11_-_26593649 | 0.02 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr6_-_139613269 | 0.02 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr2_-_201753717 | 0.01 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr2_-_207082624 | 0.01 |
ENST00000458440.1
ENST00000437420.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr4_+_95916947 | 0.01 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr15_-_100882191 | 0.01 |
ENST00000268070.4
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr21_+_17442799 | 0.01 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr16_+_57844549 | 0.01 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr4_+_26862400 | 0.01 |
ENST00000467011.1
ENST00000412829.2 |
STIM2
|
stromal interaction molecule 2 |
| chr4_-_109684120 | 0.01 |
ENST00000512646.1
ENST00000411864.2 ENST00000296486.3 ENST00000510706.1 |
ETNPPL
|
ethanolamine-phosphate phospho-lyase |
| chr18_+_32556892 | 0.01 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_-_151217001 | 0.01 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr4_-_119759795 | 0.01 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr20_+_18447771 | 0.01 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr8_+_40010989 | 0.01 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr2_+_219135115 | 0.01 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr2_-_99279928 | 0.01 |
ENST00000414521.2
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chrX_-_19988382 | 0.01 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr6_-_32157947 | 0.01 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr11_-_33913708 | 0.01 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_-_94586651 | 0.01 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr18_+_32073253 | 0.01 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr9_+_125132803 | 0.01 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_+_95027772 | 0.01 |
ENST00000555095.1
ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4
SERPINA5
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr9_+_131062367 | 0.01 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chrX_+_123097014 | 0.01 |
ENST00000394478.1
|
STAG2
|
stromal antigen 2 |
| chr7_+_129984630 | 0.01 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr18_-_48351743 | 0.01 |
ENST00000588444.1
ENST00000256425.2 ENST00000428869.2 |
MRO
|
maestro |
| chr5_-_169407744 | 0.01 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr4_+_41614909 | 0.01 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr3_-_47950745 | 0.01 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr7_-_92855762 | 0.01 |
ENST00000453812.2
ENST00000394468.2 |
HEPACAM2
|
HEPACAM family member 2 |
| chr12_-_10978957 | 0.01 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr2_-_201753980 | 0.01 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr11_+_65554493 | 0.01 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr2_-_214016314 | 0.01 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr6_-_111136513 | 0.01 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_-_50643664 | 0.01 |
ENST00000550592.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr20_+_30697298 | 0.01 |
ENST00000398022.2
|
TM9SF4
|
transmembrane 9 superfamily protein member 4 |
| chr6_+_52535878 | 0.01 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr4_+_74275057 | 0.01 |
ENST00000511370.1
|
ALB
|
albumin |
| chr17_-_60142609 | 0.01 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr8_-_110346614 | 0.01 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX3_EVX1_MEOX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0045726 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |