Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
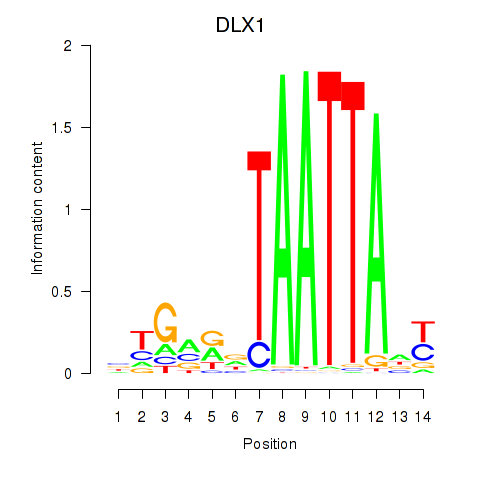
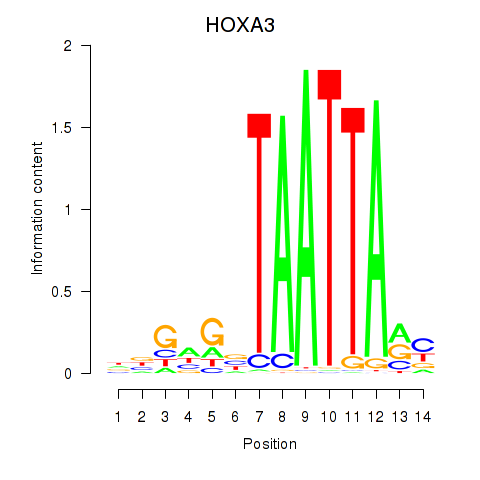
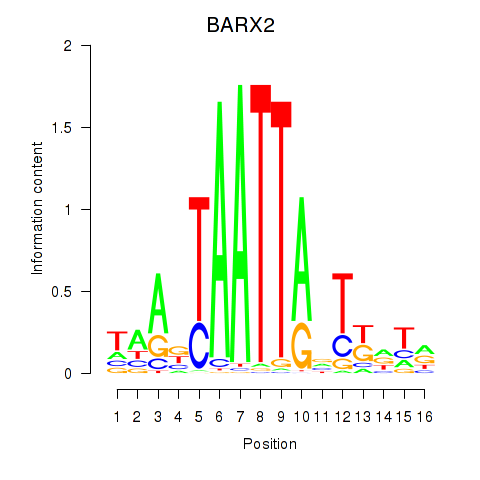
Results for DLX1_HOXA3_BARX2
Z-value: 0.63
Transcription factors associated with DLX1_HOXA3_BARX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DLX1
|
ENSG00000144355.10 | distal-less homeobox 1 |
|
HOXA3
|
ENSG00000105997.18 | homeobox A3 |
|
BARX2
|
ENSG00000043039.5 | BARX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DLX1 | hg19_v2_chr2_+_172950227_172950264 | 0.47 | 3.4e-01 | Click! |
| HOXA3 | hg19_v2_chr7_-_27153454_27153469 | 0.31 | 5.5e-01 | Click! |
| BARX2 | hg19_v2_chr11_+_129245796_129245835 | 0.08 | 8.8e-01 | Click! |
Activity profile of DLX1_HOXA3_BARX2 motif
Sorted Z-values of DLX1_HOXA3_BARX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_57229155 | 0.73 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr5_+_136070614 | 0.71 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr11_-_63376013 | 0.56 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr12_+_78359999 | 0.53 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr16_+_53412368 | 0.51 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr5_-_150473127 | 0.44 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_-_197115818 | 0.42 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr1_+_152784447 | 0.37 |
ENST00000360090.3
|
LCE1B
|
late cornified envelope 1B |
| chr18_-_59415987 | 0.37 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr7_+_37723336 | 0.35 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr17_+_61151306 | 0.35 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr9_-_5339873 | 0.35 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr19_+_45417921 | 0.31 |
ENST00000252491.4
ENST00000592885.1 ENST00000589781.1 |
APOC1
|
apolipoprotein C-I |
| chr11_-_60010556 | 0.31 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr9_-_5304432 | 0.30 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr7_-_99716952 | 0.30 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chrX_+_150565653 | 0.30 |
ENST00000330374.6
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr7_-_99716940 | 0.29 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_+_28410128 | 0.28 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr4_+_88896819 | 0.26 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr15_+_63682335 | 0.26 |
ENST00000559379.1
ENST00000559821.1 |
RP11-321G12.1
|
RP11-321G12.1 |
| chr1_-_21620877 | 0.26 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr19_+_40873617 | 0.25 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr15_+_35270552 | 0.25 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr6_-_32908765 | 0.25 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr5_+_148443049 | 0.24 |
ENST00000515304.1
ENST00000507318.1 |
CTC-529P8.1
|
CTC-529P8.1 |
| chr11_+_5775923 | 0.24 |
ENST00000317254.3
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4 (gene/pseudogene) |
| chr7_-_99716914 | 0.23 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr20_-_56265680 | 0.23 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr8_-_8639956 | 0.22 |
ENST00000522213.1
|
RP11-211C9.1
|
RP11-211C9.1 |
| chr16_+_53133070 | 0.22 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_103749313 | 0.22 |
ENST00000394803.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr4_-_69536346 | 0.21 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr12_+_8276433 | 0.20 |
ENST00000345999.3
ENST00000352620.3 ENST00000360500.3 |
CLEC4A
|
C-type lectin domain family 4, member A |
| chr2_+_149402989 | 0.20 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr6_+_46761118 | 0.19 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr6_+_149721495 | 0.19 |
ENST00000326669.4
|
SUMO4
|
small ubiquitin-like modifier 4 |
| chr7_-_99717463 | 0.19 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr15_+_96904487 | 0.18 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr11_-_26593779 | 0.18 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr10_-_115904361 | 0.18 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr16_-_28937027 | 0.18 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr3_+_98482175 | 0.17 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr19_+_17638059 | 0.17 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr6_-_28411241 | 0.17 |
ENST00000289788.4
|
ZSCAN23
|
zinc finger and SCAN domain containing 23 |
| chr10_+_81891416 | 0.17 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr16_-_25122735 | 0.17 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr5_+_66300464 | 0.17 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_+_48592134 | 0.17 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr13_-_30880979 | 0.16 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr6_+_88106840 | 0.16 |
ENST00000369570.4
|
C6orf164
|
chromosome 6 open reading frame 164 |
| chr4_-_79860506 | 0.16 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr5_+_66300446 | 0.16 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_92777606 | 0.16 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr16_+_58010339 | 0.16 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr3_+_51851612 | 0.15 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr5_-_139726181 | 0.15 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr12_+_64798095 | 0.15 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr2_-_203735484 | 0.15 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_-_76461249 | 0.15 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr3_+_138340049 | 0.14 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_+_191221240 | 0.13 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr15_+_41057818 | 0.13 |
ENST00000558467.1
|
GCHFR
|
GTP cyclohydrolase I feedback regulator |
| chr1_+_1846519 | 0.13 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr7_+_107224364 | 0.13 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr19_-_36606181 | 0.13 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr7_-_35013217 | 0.13 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr4_-_103749105 | 0.13 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_22852791 | 0.13 |
ENST00000413794.2
|
RP11-114G22.1
|
RP11-114G22.1 |
| chr17_-_72772425 | 0.13 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr11_+_24518723 | 0.13 |
ENST00000336930.6
ENST00000529015.1 ENST00000533227.1 |
LUZP2
|
leucine zipper protein 2 |
| chr18_-_33709268 | 0.13 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr11_+_18154059 | 0.13 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr17_-_72772462 | 0.13 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr15_+_94899183 | 0.13 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr3_-_33686925 | 0.13 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr8_-_55014018 | 0.12 |
ENST00000521352.1
|
LYPLA1
|
lysophospholipase I |
| chr21_-_19858196 | 0.12 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr7_+_134528635 | 0.12 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chrX_+_107288280 | 0.12 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_+_67759813 | 0.12 |
ENST00000587241.1
|
AC003051.1
|
AC003051.1 |
| chr10_+_94594351 | 0.12 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr10_+_18549645 | 0.11 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_119759795 | 0.11 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr12_+_38710555 | 0.11 |
ENST00000551464.1
|
ALG10B
|
ALG10B, alpha-1,2-glucosyltransferase |
| chr19_+_50016610 | 0.11 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr9_+_125133467 | 0.11 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_7575477 | 0.11 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr8_+_105602961 | 0.11 |
ENST00000521923.1
|
RP11-127H5.1
|
Uncharacterized protein |
| chr14_-_45252031 | 0.11 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr15_+_58702742 | 0.11 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr5_+_53686658 | 0.11 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr2_+_17997763 | 0.11 |
ENST00000281047.3
|
MSGN1
|
mesogenin 1 |
| chr17_-_57158523 | 0.11 |
ENST00000581468.1
|
TRIM37
|
tripartite motif containing 37 |
| chr4_+_189321881 | 0.10 |
ENST00000512839.1
ENST00000513313.1 |
LINC01060
|
long intergenic non-protein coding RNA 1060 |
| chr7_-_86849836 | 0.10 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr9_-_128246769 | 0.10 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr11_+_89764274 | 0.10 |
ENST00000448984.1
ENST00000432771.1 |
TRIM49C
|
tripartite motif containing 49C |
| chr12_+_26348246 | 0.10 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr6_-_47445214 | 0.10 |
ENST00000604014.1
|
RP11-385F7.1
|
RP11-385F7.1 |
| chr17_-_12920907 | 0.10 |
ENST00000609757.1
ENST00000581499.2 ENST00000580504.1 |
ELAC2
|
elaC ribonuclease Z 2 |
| chr19_-_3985455 | 0.10 |
ENST00000309311.6
|
EEF2
|
eukaryotic translation elongation factor 2 |
| chr7_+_148936732 | 0.10 |
ENST00000335870.2
|
ZNF212
|
zinc finger protein 212 |
| chrX_-_23926004 | 0.10 |
ENST00000379226.4
ENST00000379220.3 |
APOO
|
apolipoprotein O |
| chr11_-_64684672 | 0.10 |
ENST00000377264.3
ENST00000421419.2 |
ATG2A
|
autophagy related 2A |
| chr19_-_3557570 | 0.10 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_+_56814968 | 0.10 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr10_+_6779326 | 0.10 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr11_+_59705928 | 0.10 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr12_-_48164812 | 0.10 |
ENST00000549151.1
ENST00000548919.1 |
RAPGEF3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr8_-_93978309 | 0.10 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr6_+_134273321 | 0.10 |
ENST00000457715.1
|
TBPL1
|
TBP-like 1 |
| chr19_+_21579908 | 0.10 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr19_+_23257988 | 0.10 |
ENST00000594318.1
ENST00000600223.1 ENST00000593635.1 |
CTD-2291D10.4
ZNF730
|
CTD-2291D10.4 zinc finger protein 730 |
| chr16_-_1464688 | 0.10 |
ENST00000389221.4
ENST00000508903.2 ENST00000397462.1 ENST00000301712.5 |
UNKL
|
unkempt family zinc finger-like |
| chr6_+_134273300 | 0.10 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr19_+_21579958 | 0.10 |
ENST00000339914.6
ENST00000599461.1 |
ZNF493
|
zinc finger protein 493 |
| chr4_+_80584903 | 0.10 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr4_-_103749205 | 0.10 |
ENST00000508249.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_55620433 | 0.09 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr1_-_222763101 | 0.09 |
ENST00000391883.2
ENST00000366890.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr16_+_12059091 | 0.09 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_+_43885799 | 0.09 |
ENST00000449946.1
ENST00000417289.1 |
CKMT1B
|
creatine kinase, mitochondrial 1B |
| chr6_-_82957433 | 0.09 |
ENST00000306270.7
|
IBTK
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr9_-_100684769 | 0.09 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chr6_-_116833500 | 0.09 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chrX_-_115594160 | 0.09 |
ENST00000371894.4
|
CXorf61
|
cancer/testis antigen 83 |
| chr9_-_103115135 | 0.09 |
ENST00000537512.1
|
TEX10
|
testis expressed 10 |
| chr19_+_49199209 | 0.09 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr7_+_150020363 | 0.09 |
ENST00000359623.4
ENST00000493307.1 |
LRRC61
|
leucine rich repeat containing 61 |
| chr3_+_138340067 | 0.09 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_-_112993808 | 0.09 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr1_+_149239529 | 0.09 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr21_+_33671160 | 0.09 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr3_-_33686743 | 0.09 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_+_26402465 | 0.09 |
ENST00000476549.2
ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1
|
butyrophilin, subfamily 3, member A1 |
| chr17_-_15501932 | 0.09 |
ENST00000583965.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr21_-_35796241 | 0.09 |
ENST00000450895.1
|
AP000322.53
|
AP000322.53 |
| chr6_-_133035185 | 0.09 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr2_-_165630264 | 0.09 |
ENST00000452626.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr8_-_93978333 | 0.09 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr7_+_138915102 | 0.09 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr7_-_64023441 | 0.09 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr16_+_28889801 | 0.09 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr12_-_719573 | 0.09 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr4_-_175041663 | 0.08 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr11_-_327537 | 0.08 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr3_-_98235962 | 0.08 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chrX_-_20236970 | 0.08 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr8_+_94752349 | 0.08 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr14_-_39639523 | 0.08 |
ENST00000330149.5
ENST00000554018.1 ENST00000347691.5 |
TRAPPC6B
|
trafficking protein particle complex 6B |
| chr16_-_3350614 | 0.08 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr2_-_201753859 | 0.08 |
ENST00000409361.1
ENST00000392283.4 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr5_+_174151536 | 0.08 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr9_+_20927764 | 0.08 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr9_-_115095229 | 0.08 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr19_+_36606354 | 0.08 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr19_-_22018966 | 0.08 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr12_-_118628315 | 0.08 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr14_-_23426231 | 0.08 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chrX_-_9734004 | 0.08 |
ENST00000467482.1
ENST00000380929.2 |
GPR143
|
G protein-coupled receptor 143 |
| chr2_+_68592305 | 0.08 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr12_-_8088773 | 0.08 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr14_+_57671888 | 0.08 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr12_-_10282742 | 0.08 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr10_-_101825151 | 0.08 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr7_+_30589829 | 0.08 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr2_-_201753717 | 0.08 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr14_+_31046959 | 0.07 |
ENST00000547532.1
ENST00000555429.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr3_+_136649311 | 0.07 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr4_-_146261573 | 0.07 |
ENST00000610239.1
|
RP11-142A22.4
|
RP11-142A22.4 |
| chr18_+_46065393 | 0.07 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr3_+_111718173 | 0.07 |
ENST00000494932.1
|
TAGLN3
|
transgelin 3 |
| chr12_-_8088871 | 0.07 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr9_+_130911723 | 0.07 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr6_+_53964336 | 0.07 |
ENST00000447836.2
ENST00000511678.1 |
MLIP
|
muscular LMNA-interacting protein |
| chr4_-_39979576 | 0.07 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr2_+_228736335 | 0.07 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr6_+_131958436 | 0.07 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr19_+_9203855 | 0.07 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr11_+_110001723 | 0.07 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr10_+_57358750 | 0.07 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr18_-_67624160 | 0.07 |
ENST00000581982.1
ENST00000280200.4 |
CD226
|
CD226 molecule |
| chr19_+_50016411 | 0.07 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_-_143567262 | 0.07 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr1_-_159832438 | 0.07 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr18_+_55888767 | 0.07 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr17_+_73539339 | 0.07 |
ENST00000581713.1
|
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr5_+_44809027 | 0.07 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr1_+_43613612 | 0.07 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr2_+_113816215 | 0.07 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr12_-_14924056 | 0.07 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chr15_-_44969022 | 0.06 |
ENST00000560110.1
|
PATL2
|
protein associated with topoisomerase II homolog 2 (yeast) |
| chr5_+_179135246 | 0.06 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr14_+_39583427 | 0.06 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr1_-_160924589 | 0.06 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr16_+_15489629 | 0.06 |
ENST00000396385.3
|
MPV17L
|
MPV17 mitochondrial membrane protein-like |
| chr3_+_108541545 | 0.06 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr8_-_623547 | 0.06 |
ENST00000522893.1
|
ERICH1
|
glutamate-rich 1 |
| chr19_+_11071546 | 0.06 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DLX1_HOXA3_BARX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.4 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.3 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.2 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.0 | 0.3 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.0 | GO:2000625 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.0 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:0050777 | negative regulation of immune response(GO:0050777) |
| 0.0 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:0030522 | intracellular receptor signaling pathway(GO:0030522) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:1904879 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.0 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.0 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 1.2 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0036028 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.2 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0052829 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0035643 | L-DOPA receptor activity(GO:0035643) L-DOPA binding(GO:0072544) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.0 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |