Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
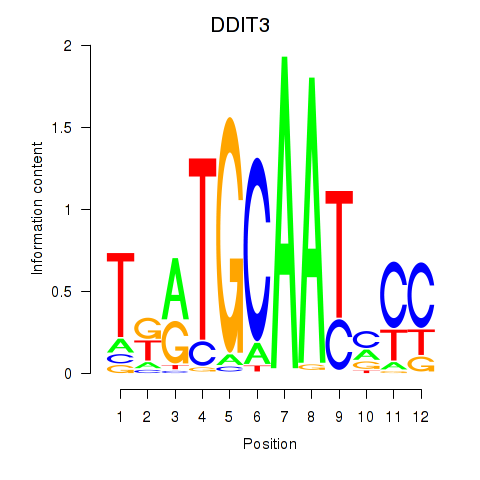
Results for DDIT3
Z-value: 0.75
Transcription factors associated with DDIT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DDIT3
|
ENSG00000175197.6 | DNA damage inducible transcript 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DDIT3 | hg19_v2_chr12_-_57914275_57914304 | 0.85 | 3.2e-02 | Click! |
Activity profile of DDIT3 motif
Sorted Z-values of DDIT3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_77967433 | 0.78 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr6_+_144606817 | 0.64 |
ENST00000433557.1
|
UTRN
|
utrophin |
| chr1_+_149804218 | 0.37 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr14_+_64565442 | 0.35 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr10_+_86004802 | 0.31 |
ENST00000359452.4
ENST00000358110.5 ENST00000372092.3 |
RGR
|
retinal G protein coupled receptor |
| chr10_-_116444371 | 0.30 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr4_+_141178440 | 0.29 |
ENST00000394205.3
|
SCOC
|
short coiled-coil protein |
| chr19_+_41497178 | 0.28 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr2_-_63815628 | 0.26 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr7_-_56101826 | 0.26 |
ENST00000421626.1
|
PSPH
|
phosphoserine phosphatase |
| chr1_+_174844645 | 0.26 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_-_86353510 | 0.25 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_-_67519782 | 0.25 |
ENST00000235345.5
|
SLC35D1
|
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr15_+_85923856 | 0.22 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr1_-_149832704 | 0.22 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr20_+_5987890 | 0.22 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr7_+_90338547 | 0.20 |
ENST00000446790.1
|
CDK14
|
cyclin-dependent kinase 14 |
| chr4_+_157997273 | 0.20 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr14_-_23446003 | 0.18 |
ENST00000553911.1
|
AJUBA
|
ajuba LIM protein |
| chr12_-_25102252 | 0.17 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr9_+_80912059 | 0.17 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr4_+_6784358 | 0.17 |
ENST00000508423.1
|
KIAA0232
|
KIAA0232 |
| chr12_-_63328817 | 0.17 |
ENST00000228705.6
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr3_-_45883558 | 0.17 |
ENST00000445698.1
ENST00000296135.6 |
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr15_-_83837983 | 0.17 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr15_-_64126084 | 0.16 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr15_+_85923797 | 0.16 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr5_+_112074029 | 0.16 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr19_-_6604094 | 0.16 |
ENST00000597430.2
|
CD70
|
CD70 molecule |
| chr14_+_62462541 | 0.16 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr6_+_80341000 | 0.15 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr16_+_56970567 | 0.15 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr12_+_64798826 | 0.15 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr4_-_99578789 | 0.15 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr18_-_55288973 | 0.15 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr3_+_38017264 | 0.15 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr7_-_130597935 | 0.14 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr12_+_32638897 | 0.14 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_63816295 | 0.13 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr2_+_63816126 | 0.13 |
ENST00000454035.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr2_+_63816087 | 0.13 |
ENST00000409908.1
ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr3_+_136676707 | 0.12 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr12_-_25101920 | 0.12 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr3_+_136676851 | 0.12 |
ENST00000309741.5
|
IL20RB
|
interleukin 20 receptor beta |
| chr5_-_137878887 | 0.12 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr14_+_32798547 | 0.12 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_92681033 | 0.11 |
ENST00000371697.3
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle) |
| chr11_-_8680383 | 0.11 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr2_-_208030886 | 0.10 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr14_-_23446021 | 0.10 |
ENST00000553592.1
|
AJUBA
|
ajuba LIM protein |
| chr2_-_43453734 | 0.10 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr16_+_56965960 | 0.10 |
ENST00000439977.2
ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr11_+_47279155 | 0.10 |
ENST00000444396.1
ENST00000457932.1 ENST00000412937.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr7_-_130598059 | 0.09 |
ENST00000432045.2
|
MIR29B1
|
microRNA 29a |
| chr4_+_6784401 | 0.09 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr5_+_72143988 | 0.09 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr11_+_34073195 | 0.09 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr2_-_101925055 | 0.09 |
ENST00000295317.3
|
RNF149
|
ring finger protein 149 |
| chrX_-_138724677 | 0.09 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr11_-_10315741 | 0.09 |
ENST00000256190.8
|
SBF2
|
SET binding factor 2 |
| chr2_+_63816269 | 0.08 |
ENST00000432309.1
|
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr10_+_12391481 | 0.08 |
ENST00000378847.3
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr12_+_50144381 | 0.08 |
ENST00000552370.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_-_184243561 | 0.08 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr9_-_124991124 | 0.08 |
ENST00000394319.4
ENST00000340587.3 |
LHX6
|
LIM homeobox 6 |
| chr11_-_104972158 | 0.08 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr8_+_26435915 | 0.07 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr12_-_122017542 | 0.07 |
ENST00000446152.2
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr14_-_92413727 | 0.07 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr14_-_74226961 | 0.07 |
ENST00000286523.5
ENST00000435371.1 |
ELMSAN1
|
ELM2 and Myb/SANT-like domain containing 1 |
| chr11_+_62495541 | 0.07 |
ENST00000530625.1
ENST00000513247.2 |
TTC9C
|
tetratricopeptide repeat domain 9C |
| chr11_-_33913708 | 0.07 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr1_-_114447412 | 0.07 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr20_-_60294804 | 0.06 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr21_-_46334186 | 0.06 |
ENST00000522931.1
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_+_111770232 | 0.06 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr7_+_99006232 | 0.06 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr3_+_130569429 | 0.06 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr3_+_15643476 | 0.06 |
ENST00000436193.1
ENST00000383778.4 |
BTD
|
biotinidase |
| chr2_-_161350305 | 0.06 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_+_44840679 | 0.05 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr1_-_152196669 | 0.05 |
ENST00000368801.2
|
HRNR
|
hornerin |
| chr8_+_27629459 | 0.05 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr9_+_110045537 | 0.05 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr14_+_32798462 | 0.05 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr3_-_148939835 | 0.05 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr5_+_145583156 | 0.05 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr17_-_46691990 | 0.04 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr17_-_30228678 | 0.04 |
ENST00000261708.4
|
UTP6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr20_+_54987168 | 0.04 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr9_-_95055956 | 0.04 |
ENST00000375629.3
ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS
|
isoleucyl-tRNA synthetase |
| chr3_+_186330712 | 0.04 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr20_+_24929866 | 0.04 |
ENST00000480798.1
ENST00000376835.2 |
CST7
|
cystatin F (leukocystatin) |
| chr19_-_49258606 | 0.04 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr5_+_145583107 | 0.04 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr11_+_47279504 | 0.04 |
ENST00000441012.2
ENST00000437276.1 ENST00000436029.1 ENST00000467728.1 ENST00000405853.3 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr16_-_18908196 | 0.04 |
ENST00000565324.1
ENST00000561947.1 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr11_+_34073269 | 0.04 |
ENST00000389645.3
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr17_-_2117600 | 0.03 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr2_-_208030647 | 0.03 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr6_+_151187074 | 0.03 |
ENST00000367308.4
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr13_-_45915221 | 0.03 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr12_-_12491608 | 0.03 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr9_-_95056010 | 0.03 |
ENST00000443024.2
|
IARS
|
isoleucyl-tRNA synthetase |
| chr3_+_12392971 | 0.03 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr8_-_18744528 | 0.03 |
ENST00000523619.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr13_-_24007815 | 0.03 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr4_+_111397216 | 0.03 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr17_-_34257771 | 0.02 |
ENST00000394529.3
ENST00000293273.6 |
RDM1
|
RAD52 motif 1 |
| chr7_-_99006443 | 0.02 |
ENST00000350498.3
|
PDAP1
|
PDGFA associated protein 1 |
| chr1_+_114447763 | 0.02 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr22_+_31518938 | 0.02 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr1_+_111770278 | 0.02 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr9_-_77502636 | 0.02 |
ENST00000449912.2
|
TRPM6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr1_-_207143802 | 0.02 |
ENST00000324852.4
ENST00000400962.3 |
FCAMR
|
Fc receptor, IgA, IgM, high affinity |
| chr1_+_150954493 | 0.02 |
ENST00000368947.4
|
ANXA9
|
annexin A9 |
| chr19_+_33865218 | 0.02 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr19_-_35992780 | 0.02 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr2_+_68872954 | 0.02 |
ENST00000394342.2
|
PROKR1
|
prokineticin receptor 1 |
| chr10_+_103986085 | 0.02 |
ENST00000370005.3
|
ELOVL3
|
ELOVL fatty acid elongase 3 |
| chr2_+_227700652 | 0.02 |
ENST00000341329.3
ENST00000392062.2 ENST00000437454.1 ENST00000443477.1 ENST00000423616.1 ENST00000448992.1 |
RHBDD1
|
rhomboid domain containing 1 |
| chr6_+_31895287 | 0.02 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr12_+_57857475 | 0.01 |
ENST00000528467.1
|
GLI1
|
GLI family zinc finger 1 |
| chr10_+_60028818 | 0.01 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr5_-_131630931 | 0.01 |
ENST00000431054.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr9_+_112542591 | 0.01 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr13_+_24844979 | 0.01 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chrX_-_138724994 | 0.00 |
ENST00000536274.1
|
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr1_+_90308981 | 0.00 |
ENST00000527156.1
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr10_+_102756800 | 0.00 |
ENST00000370223.3
|
LZTS2
|
leucine zipper, putative tumor suppressor 2 |
| chr10_-_13523073 | 0.00 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr19_+_12917364 | 0.00 |
ENST00000221486.4
|
RNASEH2A
|
ribonuclease H2, subunit A |
| chr11_+_62495997 | 0.00 |
ENST00000316461.4
|
TTC9C
|
tetratricopeptide repeat domain 9C |
Network of associatons between targets according to the STRING database.
First level regulatory network of DDIT3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.1 | 0.2 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.3 | GO:0009099 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.4 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.2 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.1 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.1 | 0.2 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.3 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |