Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
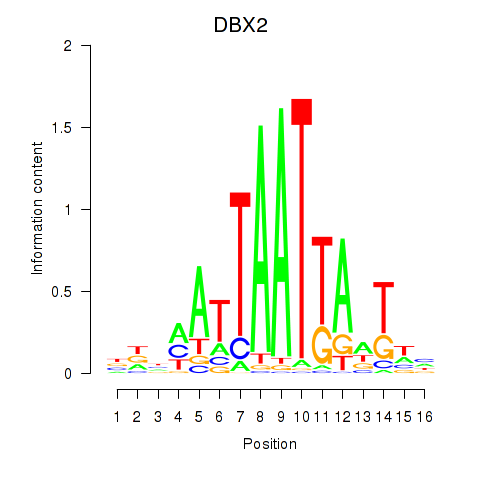
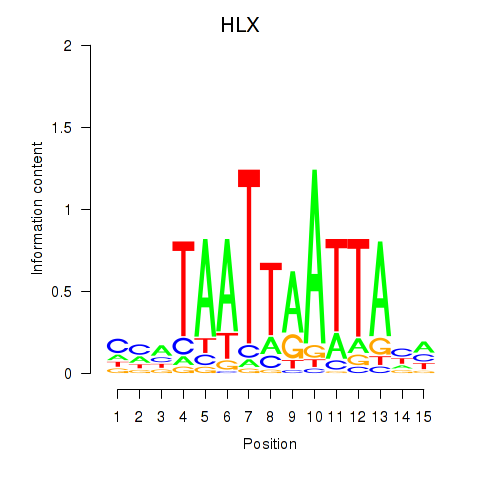
Results for DBX2_HLX
Z-value: 0.60
Transcription factors associated with DBX2_HLX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBX2
|
ENSG00000185610.6 | developing brain homeobox 2 |
|
HLX
|
ENSG00000136630.11 | H2.0 like homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HLX | hg19_v2_chr1_+_221054584_221054584 | 0.75 | 8.5e-02 | Click! |
Activity profile of DBX2_HLX motif
Sorted Z-values of DBX2_HLX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_35013217 | 0.56 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr4_-_89442940 | 0.45 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chrX_+_3189861 | 0.43 |
ENST00000457435.1
ENST00000420429.2 |
CXorf28
|
chromosome X open reading frame 28 |
| chr9_-_95055923 | 0.41 |
ENST00000430417.1
|
IARS
|
isoleucyl-tRNA synthetase |
| chr9_+_67977438 | 0.40 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr16_+_53412368 | 0.37 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr12_+_41136144 | 0.37 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr7_-_64023441 | 0.37 |
ENST00000309683.6
|
ZNF680
|
zinc finger protein 680 |
| chr22_-_29107919 | 0.37 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr11_-_26593779 | 0.32 |
ENST00000529533.1
|
MUC15
|
mucin 15, cell surface associated |
| chr9_-_21482312 | 0.31 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr1_+_117963209 | 0.31 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr13_-_86373536 | 0.30 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chrX_+_77166172 | 0.30 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr2_+_161993465 | 0.30 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr1_-_168464875 | 0.29 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr3_+_38537960 | 0.28 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr5_+_67588312 | 0.27 |
ENST00000519025.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_100356551 | 0.27 |
ENST00000209665.4
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr17_-_8263538 | 0.26 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr4_+_95128748 | 0.25 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_-_112993808 | 0.24 |
ENST00000511219.1
|
RP11-269F21.3
|
RP11-269F21.3 |
| chr8_+_42396274 | 0.24 |
ENST00000438528.3
|
SMIM19
|
small integral membrane protein 19 |
| chr2_+_172309634 | 0.24 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr14_-_36990061 | 0.24 |
ENST00000546983.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr5_+_147691979 | 0.24 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chr8_-_27695552 | 0.24 |
ENST00000522944.1
ENST00000301905.4 |
PBK
|
PDZ binding kinase |
| chr7_+_13141010 | 0.24 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr4_-_100356291 | 0.24 |
ENST00000476959.1
ENST00000482593.1 |
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_-_52859046 | 0.23 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr16_-_3350614 | 0.23 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr5_+_102200948 | 0.23 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_-_84122033 | 0.23 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr9_-_3469181 | 0.23 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr12_-_10573149 | 0.23 |
ENST00000381904.2
ENST00000381903.2 ENST00000396439.2 |
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3 |
| chr2_-_17981462 | 0.22 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr10_-_115904361 | 0.22 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr12_-_10282742 | 0.22 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr7_+_116654958 | 0.21 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr4_-_118006697 | 0.20 |
ENST00000310754.4
|
TRAM1L1
|
translocation associated membrane protein 1-like 1 |
| chr5_+_115177178 | 0.20 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr16_-_28937027 | 0.20 |
ENST00000358201.4
|
RABEP2
|
rabaptin, RAB GTPase binding effector protein 2 |
| chr11_-_26593677 | 0.19 |
ENST00000527569.1
|
MUC15
|
mucin 15, cell surface associated |
| chrX_+_150565038 | 0.19 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr1_-_67142710 | 0.19 |
ENST00000502413.2
|
AL139147.1
|
Uncharacterized protein |
| chr18_+_61445205 | 0.19 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_224544572 | 0.19 |
ENST00000366857.5
ENST00000366856.3 |
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr9_+_125133315 | 0.18 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_-_42358742 | 0.18 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr1_+_234765057 | 0.18 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr1_+_76251912 | 0.18 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr2_-_98972468 | 0.18 |
ENST00000454230.1
|
AC092675.3
|
Uncharacterized protein |
| chr11_-_559377 | 0.17 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr1_-_54411255 | 0.17 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr8_-_71157595 | 0.17 |
ENST00000519724.1
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr17_+_17685422 | 0.17 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr1_+_84630574 | 0.17 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr19_-_22018966 | 0.17 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr12_-_10022735 | 0.17 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr14_+_55494323 | 0.17 |
ENST00000339298.2
|
SOCS4
|
suppressor of cytokine signaling 4 |
| chr10_-_28571015 | 0.17 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr9_+_125132803 | 0.17 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr12_-_76461249 | 0.17 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_+_81771806 | 0.16 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr1_-_160549235 | 0.16 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr4_+_75174180 | 0.16 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr1_+_152730499 | 0.16 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chrX_-_73072534 | 0.16 |
ENST00000429829.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr15_+_75970672 | 0.16 |
ENST00000435356.1
|
AC105020.1
|
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr4_+_74301880 | 0.16 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr6_+_153552455 | 0.16 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr3_+_138340067 | 0.16 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr2_+_183982255 | 0.15 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr14_+_55493920 | 0.15 |
ENST00000395472.2
ENST00000555846.1 |
SOCS4
|
suppressor of cytokine signaling 4 |
| chr16_-_47493041 | 0.15 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr6_-_52859968 | 0.15 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_+_103089756 | 0.15 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr6_+_114178512 | 0.14 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr9_+_125133467 | 0.14 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_+_9296279 | 0.14 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr12_+_64798095 | 0.14 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr15_-_75748143 | 0.14 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr15_+_92006567 | 0.14 |
ENST00000554333.1
|
RP11-661P17.1
|
RP11-661P17.1 |
| chr17_+_68047418 | 0.14 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr7_+_129932974 | 0.13 |
ENST00000445470.2
ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4
|
carboxypeptidase A4 |
| chr2_+_211342400 | 0.13 |
ENST00000417946.1
ENST00000518043.1 ENST00000523702.1 |
CPS1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| chr5_+_179135246 | 0.13 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr11_+_100784231 | 0.13 |
ENST00000531183.1
|
ARHGAP42
|
Rho GTPase activating protein 42 |
| chr20_-_55934878 | 0.13 |
ENST00000543500.1
|
MTRNR2L3
|
MT-RNR2-like 3 |
| chr8_-_42396185 | 0.13 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr2_+_201242941 | 0.13 |
ENST00000449647.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr12_+_80838126 | 0.13 |
ENST00000266688.5
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q |
| chr10_-_99030395 | 0.13 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr1_-_204183071 | 0.12 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr1_-_186365908 | 0.12 |
ENST00000598663.1
|
AL596220.1
|
Uncharacterized protein |
| chr5_+_67588391 | 0.12 |
ENST00000523872.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_160370344 | 0.12 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr2_+_120687335 | 0.12 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr12_+_147052 | 0.12 |
ENST00000594563.1
|
AC026369.1
|
Uncharacterized protein |
| chr6_+_26087509 | 0.12 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr9_-_5833027 | 0.12 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr11_-_124670550 | 0.12 |
ENST00000239614.4
|
MSANTD2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr7_-_99716914 | 0.12 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr2_+_109204909 | 0.12 |
ENST00000393310.1
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_29001711 | 0.12 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr7_-_140482926 | 0.12 |
ENST00000496384.2
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr1_+_84630645 | 0.12 |
ENST00000394839.2
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr18_+_48918368 | 0.12 |
ENST00000583982.1
ENST00000578152.1 ENST00000583609.1 ENST00000435144.1 ENST00000580841.1 |
RP11-267C16.1
|
RP11-267C16.1 |
| chr3_-_20053741 | 0.12 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr3_+_82035289 | 0.12 |
ENST00000470263.1
ENST00000494340.1 |
RP11-260O18.1
|
RP11-260O18.1 |
| chr12_-_76817036 | 0.11 |
ENST00000546946.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr5_+_54455946 | 0.11 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr5_+_95066823 | 0.11 |
ENST00000506817.1
ENST00000379982.3 |
RHOBTB3
|
Rho-related BTB domain containing 3 |
| chr19_-_44388116 | 0.11 |
ENST00000587539.1
|
ZNF404
|
zinc finger protein 404 |
| chr8_-_133637624 | 0.11 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr2_+_191221240 | 0.11 |
ENST00000409027.1
ENST00000458193.1 |
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr20_+_19738300 | 0.11 |
ENST00000432334.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chr2_+_149402989 | 0.11 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr16_-_28634874 | 0.11 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr14_-_92247032 | 0.11 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr4_+_159131630 | 0.11 |
ENST00000504569.1
ENST00000509278.1 ENST00000514558.1 ENST00000503200.1 |
TMEM144
|
transmembrane protein 144 |
| chr19_-_3557570 | 0.11 |
ENST00000355415.2
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr4_+_56815102 | 0.11 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr2_-_87248975 | 0.11 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr11_-_111649074 | 0.11 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr4_+_41540160 | 0.11 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_152779104 | 0.11 |
ENST00000606576.1
ENST00000368768.1 |
LCE1C
|
late cornified envelope 1C |
| chr17_+_1674982 | 0.10 |
ENST00000572048.1
ENST00000573763.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr13_+_41363581 | 0.10 |
ENST00000338625.4
|
SLC25A15
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
| chr8_+_107630340 | 0.10 |
ENST00000497705.1
|
OXR1
|
oxidation resistance 1 |
| chr10_+_91061712 | 0.10 |
ENST00000371826.3
|
IFIT2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr19_-_19739321 | 0.10 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr14_+_60558627 | 0.10 |
ENST00000317623.4
ENST00000391611.2 ENST00000406854.1 ENST00000406949.1 |
PCNXL4
|
pecanex-like 4 (Drosophila) |
| chr4_+_139694701 | 0.10 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chr15_+_66585950 | 0.10 |
ENST00000525109.1
|
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr1_-_154178803 | 0.10 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr17_-_60142609 | 0.10 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr2_+_87135076 | 0.10 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chrX_-_138724677 | 0.10 |
ENST00000370573.4
ENST00000338585.6 ENST00000370576.4 |
MCF2
|
MCF.2 cell line derived transforming sequence |
| chr11_+_17281900 | 0.10 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr4_-_185275104 | 0.10 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr1_+_62439037 | 0.10 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr19_+_50016610 | 0.10 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_+_67498538 | 0.10 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr16_+_24549014 | 0.10 |
ENST00000564314.1
ENST00000567686.1 |
RBBP6
|
retinoblastoma binding protein 6 |
| chr3_+_44916098 | 0.10 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr4_+_48833183 | 0.10 |
ENST00000503016.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr13_-_45048386 | 0.09 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr4_+_123161120 | 0.09 |
ENST00000446180.1
|
KIAA1109
|
KIAA1109 |
| chr10_-_74283694 | 0.09 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr1_-_108743471 | 0.09 |
ENST00000569674.1
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr12_-_8218997 | 0.09 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr7_-_22862406 | 0.09 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr7_+_39773160 | 0.09 |
ENST00000340510.4
|
LINC00265
|
long intergenic non-protein coding RNA 265 |
| chr16_+_14802801 | 0.09 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr19_+_42212526 | 0.09 |
ENST00000598976.1
ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA
CEACAM5
|
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr1_+_196788887 | 0.09 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chrX_-_30871004 | 0.09 |
ENST00000378928.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr1_+_212475148 | 0.09 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr22_+_46476192 | 0.09 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr19_+_21579908 | 0.09 |
ENST00000596302.1
ENST00000392288.2 ENST00000594390.1 ENST00000355504.4 |
ZNF493
|
zinc finger protein 493 |
| chr12_-_100656134 | 0.09 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr4_+_186990298 | 0.09 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr7_-_7575477 | 0.09 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr11_-_26593649 | 0.09 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr8_-_54752406 | 0.09 |
ENST00000520188.1
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chrX_-_77225135 | 0.09 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr10_-_116444371 | 0.09 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr5_+_68860949 | 0.09 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr11_-_7950209 | 0.08 |
ENST00000309838.2
|
OR10A6
|
olfactory receptor, family 10, subfamily A, member 6 |
| chr6_+_55192267 | 0.08 |
ENST00000340465.2
|
GFRAL
|
GDNF family receptor alpha like |
| chr4_+_109571740 | 0.08 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr17_+_29664830 | 0.08 |
ENST00000444181.2
ENST00000417592.2 |
NF1
|
neurofibromin 1 |
| chr8_+_120885949 | 0.08 |
ENST00000523492.1
ENST00000286234.5 |
DEPTOR
|
DEP domain containing MTOR-interacting protein |
| chr2_+_29320571 | 0.08 |
ENST00000401605.1
ENST00000401617.2 |
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr16_+_15489603 | 0.08 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chrX_-_73061339 | 0.08 |
ENST00000602863.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr3_-_123512688 | 0.08 |
ENST00000475616.1
|
MYLK
|
myosin light chain kinase |
| chr8_+_62200509 | 0.08 |
ENST00000519846.1
ENST00000518592.1 ENST00000325897.4 |
CLVS1
|
clavesin 1 |
| chr2_+_169312350 | 0.08 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr5_+_136070614 | 0.08 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr5_-_74162739 | 0.08 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chrX_-_131547596 | 0.08 |
ENST00000538204.1
ENST00000370849.3 |
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr22_-_24126145 | 0.08 |
ENST00000598975.1
|
AP000349.1
|
Uncharacterized protein |
| chr3_-_196911002 | 0.08 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr9_+_34652164 | 0.08 |
ENST00000441545.2
ENST00000553620.1 |
IL11RA
|
interleukin 11 receptor, alpha |
| chr6_-_119031228 | 0.08 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chrX_+_56590002 | 0.08 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr12_+_28605426 | 0.07 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr5_+_43602750 | 0.07 |
ENST00000505678.2
ENST00000512422.1 ENST00000264663.5 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_-_145754428 | 0.07 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr3_-_108248169 | 0.07 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr14_-_53258314 | 0.07 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr3_+_195447738 | 0.07 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr12_-_118796910 | 0.07 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr12_+_122688090 | 0.07 |
ENST00000324189.4
ENST00000546192.1 |
B3GNT4
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr15_+_75639372 | 0.07 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr4_-_74486109 | 0.07 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr9_-_27005686 | 0.07 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr12_-_46121554 | 0.07 |
ENST00000609803.1
|
LINC00938
|
long intergenic non-protein coding RNA 938 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBX2_HLX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.4 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.1 | 0.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.2 | GO:1904017 | cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.6 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:0070409 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.1 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.6 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.2 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.5 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.0 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.0 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.0 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.3 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.2 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.0 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |