Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
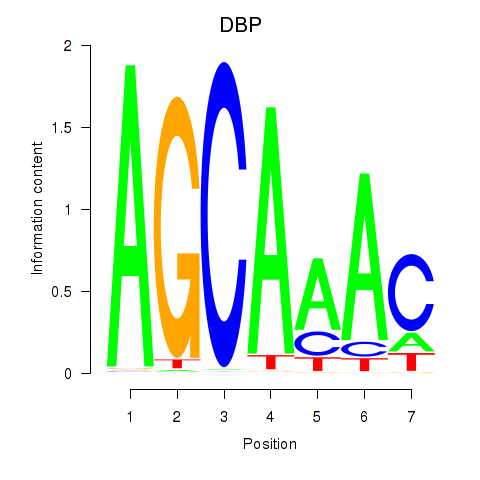
Results for DBP
Z-value: 1.02
Transcription factors associated with DBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DBP
|
ENSG00000105516.6 | D-box binding PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DBP | hg19_v2_chr19_-_49140692_49140709 | 0.61 | 2.0e-01 | Click! |
Activity profile of DBP motif
Sorted Z-values of DBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_17791648 | 1.14 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr4_-_23891693 | 0.81 |
ENST00000264867.2
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr17_+_26833250 | 0.71 |
ENST00000577936.1
ENST00000579795.1 |
FOXN1
|
forkhead box N1 |
| chr8_-_72268721 | 0.40 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr13_+_97874574 | 0.39 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr16_-_79634595 | 0.38 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr2_-_164592497 | 0.38 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr1_-_108231101 | 0.38 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chr17_-_8263538 | 0.38 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr8_+_86999516 | 0.38 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr13_-_41593425 | 0.36 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr17_+_67498396 | 0.36 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr4_-_184243561 | 0.36 |
ENST00000514470.1
ENST00000541814.1 |
CLDN24
|
claudin 24 |
| chr21_+_17791838 | 0.32 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr13_+_97928395 | 0.31 |
ENST00000445661.2
|
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr11_-_47206965 | 0.30 |
ENST00000525725.1
|
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_+_394145 | 0.29 |
ENST00000528036.1
|
PKP3
|
plakophilin 3 |
| chr1_-_40367668 | 0.29 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chrX_-_83442915 | 0.29 |
ENST00000262752.2
ENST00000543399.1 |
RPS6KA6
|
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr9_+_2017063 | 0.29 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr14_-_73360796 | 0.27 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr8_+_134125727 | 0.27 |
ENST00000521107.1
|
TG
|
thyroglobulin |
| chr9_+_2158443 | 0.27 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_61547405 | 0.27 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr6_-_31514516 | 0.26 |
ENST00000303892.5
ENST00000483251.1 |
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr10_-_81203972 | 0.26 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr14_-_23652849 | 0.26 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr22_-_31742218 | 0.26 |
ENST00000266269.5
ENST00000405309.3 ENST00000351933.4 |
PATZ1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr6_+_90272488 | 0.26 |
ENST00000485637.1
ENST00000522705.1 |
ANKRD6
|
ankyrin repeat domain 6 |
| chr8_-_102803163 | 0.25 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr9_+_2158485 | 0.25 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr11_+_121461097 | 0.24 |
ENST00000527934.1
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chrX_+_28605516 | 0.24 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr6_+_26045603 | 0.24 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chrX_+_10031499 | 0.23 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr4_-_100242549 | 0.22 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chrX_-_111923145 | 0.22 |
ENST00000371968.3
ENST00000536453.1 |
LHFPL1
|
lipoma HMGIC fusion partner-like 1 |
| chr18_-_25616519 | 0.22 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr20_-_14318248 | 0.22 |
ENST00000378053.3
ENST00000341420.4 |
FLRT3
|
fibronectin leucine rich transmembrane protein 3 |
| chr1_+_206623784 | 0.21 |
ENST00000426388.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr8_-_22550815 | 0.21 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr12_-_92536433 | 0.21 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr16_+_30960375 | 0.20 |
ENST00000318663.4
ENST00000566237.1 ENST00000562699.1 |
ORAI3
|
ORAI calcium release-activated calcium modulator 3 |
| chr17_+_42429493 | 0.20 |
ENST00000586242.1
|
GRN
|
granulin |
| chr8_-_17533838 | 0.20 |
ENST00000400046.1
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr3_-_33700589 | 0.20 |
ENST00000461133.3
ENST00000496954.2 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_-_18122843 | 0.20 |
ENST00000340650.3
|
NHLRC1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr1_+_12538594 | 0.19 |
ENST00000543710.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr3_-_171489085 | 0.19 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr10_-_79397202 | 0.18 |
ENST00000372437.1
ENST00000372408.2 ENST00000372403.4 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_-_10851762 | 0.18 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr5_+_50679506 | 0.18 |
ENST00000511384.1
|
ISL1
|
ISL LIM homeobox 1 |
| chr12_+_18891045 | 0.18 |
ENST00000317658.3
|
CAPZA3
|
capping protein (actin filament) muscle Z-line, alpha 3 |
| chr1_-_151148442 | 0.18 |
ENST00000441701.1
ENST00000416280.2 |
TMOD4
|
tropomodulin 4 (muscle) |
| chrX_+_134654540 | 0.18 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chrX_+_153769446 | 0.18 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr4_+_24797085 | 0.18 |
ENST00000382120.3
|
SOD3
|
superoxide dismutase 3, extracellular |
| chr19_+_41856816 | 0.17 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr1_-_40367530 | 0.17 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chrX_-_151619746 | 0.17 |
ENST00000370314.4
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr16_+_86612112 | 0.17 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr1_+_229440129 | 0.16 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr11_-_16403910 | 0.16 |
ENST00000526673.1
|
SOX6
|
SRY (sex determining region Y)-box 6 |
| chr11_+_34999328 | 0.16 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr2_-_174830430 | 0.16 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr6_-_75953484 | 0.15 |
ENST00000472311.2
ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr2_+_234602305 | 0.15 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr5_+_180467259 | 0.15 |
ENST00000515271.1
|
BTNL9
|
butyrophilin-like 9 |
| chr2_+_29001711 | 0.15 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_-_186732048 | 0.15 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_32710736 | 0.14 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr14_+_45366518 | 0.14 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr10_-_79397316 | 0.14 |
ENST00000372421.5
ENST00000457953.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chrX_+_49832231 | 0.14 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr6_+_45296391 | 0.14 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr3_+_153202284 | 0.14 |
ENST00000446603.2
|
C3orf79
|
chromosome 3 open reading frame 79 |
| chr20_+_9146969 | 0.14 |
ENST00000416836.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr10_-_79397391 | 0.14 |
ENST00000286628.8
ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_104104379 | 0.14 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr3_+_190333097 | 0.13 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chrX_+_149531524 | 0.13 |
ENST00000370401.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chr12_-_62586543 | 0.13 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr7_+_127233689 | 0.13 |
ENST00000265825.5
ENST00000420086.2 |
FSCN3
|
fascin homolog 3, actin-bundling protein, testicular (Strongylocentrotus purpuratus) |
| chr4_-_87281196 | 0.13 |
ENST00000359221.3
|
MAPK10
|
mitogen-activated protein kinase 10 |
| chr16_+_30675654 | 0.13 |
ENST00000287468.5
ENST00000395073.2 |
FBRS
|
fibrosin |
| chr17_+_67498538 | 0.13 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr18_-_21891460 | 0.12 |
ENST00000357041.4
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr10_+_115511434 | 0.12 |
ENST00000369312.4
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr19_-_41220957 | 0.12 |
ENST00000596357.1
ENST00000243583.6 ENST00000600080.1 ENST00000595254.1 ENST00000601967.1 |
ADCK4
|
aarF domain containing kinase 4 |
| chr6_-_56819385 | 0.12 |
ENST00000370754.5
ENST00000449297.2 |
DST
|
dystonin |
| chr19_+_13135386 | 0.12 |
ENST00000360105.4
ENST00000588228.1 ENST00000591028.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr14_-_23479331 | 0.12 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr17_+_72426891 | 0.12 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_+_394196 | 0.12 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr1_+_87797351 | 0.12 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr1_+_202385953 | 0.12 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_-_6483969 | 0.12 |
ENST00000396966.2
|
SCNN1A
|
sodium channel, non-voltage-gated 1 alpha subunit |
| chrX_+_149531606 | 0.11 |
ENST00000432680.2
|
MAMLD1
|
mastermind-like domain containing 1 |
| chrX_+_16141667 | 0.11 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chr6_-_42419649 | 0.11 |
ENST00000372922.4
ENST00000541110.1 ENST00000372917.4 |
TRERF1
|
transcriptional regulating factor 1 |
| chr5_-_111091948 | 0.11 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr8_-_6420759 | 0.11 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr8_-_22549856 | 0.11 |
ENST00000522910.1
|
EGR3
|
early growth response 3 |
| chr2_+_191002486 | 0.11 |
ENST00000396974.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr18_+_61223393 | 0.11 |
ENST00000269491.1
ENST00000382768.1 |
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr10_+_115511213 | 0.11 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chrX_+_49644470 | 0.11 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr1_+_162039558 | 0.11 |
ENST00000530878.1
ENST00000361897.5 |
NOS1AP
|
nitric oxide synthase 1 (neuronal) adaptor protein |
| chr2_+_234601512 | 0.11 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr9_+_139886846 | 0.11 |
ENST00000371620.3
|
C9orf142
|
chromosome 9 open reading frame 142 |
| chr17_-_34308524 | 0.11 |
ENST00000293275.3
|
CCL16
|
chemokine (C-C motif) ligand 16 |
| chr12_+_65218352 | 0.11 |
ENST00000539867.1
ENST00000544457.1 ENST00000539120.1 |
TBC1D30
|
TBC1 domain family, member 30 |
| chr2_+_166326157 | 0.10 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr2_+_166430619 | 0.10 |
ENST00000409420.1
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr12_-_120765565 | 0.10 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chrX_+_56590002 | 0.10 |
ENST00000338222.5
|
UBQLN2
|
ubiquilin 2 |
| chr13_+_44947941 | 0.10 |
ENST00000379179.3
|
SERP2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr9_+_72658490 | 0.10 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr10_-_31288398 | 0.10 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr20_+_18447771 | 0.10 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr1_+_24645807 | 0.10 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_-_238108575 | 0.10 |
ENST00000604646.1
|
MTRNR2L11
|
MT-RNR2-like 11 (pseudogene) |
| chr1_+_24645865 | 0.10 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr10_-_99052382 | 0.10 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr14_-_36988882 | 0.10 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr4_-_87281224 | 0.09 |
ENST00000395169.3
ENST00000395161.2 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_106473768 | 0.09 |
ENST00000265154.2
ENST00000420470.2 |
ARHGEF38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr14_+_93357749 | 0.09 |
ENST00000557689.1
|
RP11-862G15.1
|
RP11-862G15.1 |
| chr3_-_112564797 | 0.09 |
ENST00000398214.1
ENST00000448932.1 |
CD200R1L
|
CD200 receptor 1-like |
| chr17_-_48546324 | 0.09 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr10_-_79397479 | 0.09 |
ENST00000404771.3
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr10_+_21823243 | 0.09 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chrX_+_129473859 | 0.09 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr10_-_64576105 | 0.09 |
ENST00000242480.3
ENST00000411732.1 |
EGR2
|
early growth response 2 |
| chr17_+_48823896 | 0.09 |
ENST00000511974.1
|
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr1_+_24645915 | 0.09 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr17_-_56492989 | 0.09 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr7_+_13141097 | 0.09 |
ENST00000411542.1
|
AC011288.2
|
AC011288.2 |
| chr3_-_52479043 | 0.08 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr1_+_24646002 | 0.08 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr18_+_52385068 | 0.08 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr5_-_124080203 | 0.08 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr8_-_101718991 | 0.08 |
ENST00000517990.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr17_+_48624450 | 0.08 |
ENST00000006658.6
ENST00000356488.4 ENST00000393244.3 |
SPATA20
|
spermatogenesis associated 20 |
| chr22_+_45714361 | 0.08 |
ENST00000452238.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr1_-_85870177 | 0.08 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr5_+_67576062 | 0.08 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_76555657 | 0.08 |
ENST00000307465.4
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase) |
| chr1_+_61548225 | 0.08 |
ENST00000371187.3
|
NFIA
|
nuclear factor I/A |
| chr7_-_150946015 | 0.08 |
ENST00000262188.8
|
SMARCD3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr8_+_107738240 | 0.08 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr10_+_104178946 | 0.08 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr1_+_84609944 | 0.08 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr9_+_35829208 | 0.08 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr20_+_11898507 | 0.08 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr19_+_39881951 | 0.07 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr1_-_23886285 | 0.07 |
ENST00000374561.5
|
ID3
|
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr17_+_28705921 | 0.07 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr1_+_203734296 | 0.07 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr7_+_26331678 | 0.07 |
ENST00000446848.2
|
SNX10
|
sorting nexin 10 |
| chr3_-_48471454 | 0.07 |
ENST00000296440.6
ENST00000448774.2 |
PLXNB1
|
plexin B1 |
| chr8_-_22550691 | 0.07 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chrX_+_49028265 | 0.07 |
ENST00000376322.3
ENST00000376327.5 |
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched) |
| chr17_-_48546232 | 0.07 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr2_-_211036051 | 0.07 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr12_-_99548524 | 0.07 |
ENST00000549558.2
ENST00000550693.2 ENST00000549493.2 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_-_89978299 | 0.07 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr2_-_111291587 | 0.07 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr14_+_32963433 | 0.07 |
ENST00000554410.1
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_27648709 | 0.07 |
ENST00000608611.1
ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222
|
transmembrane protein 222 |
| chr3_-_50541028 | 0.07 |
ENST00000266039.3
ENST00000435965.1 ENST00000395083.1 |
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr11_-_62414070 | 0.06 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chrX_-_54070607 | 0.06 |
ENST00000338154.6
ENST00000338946.6 |
PHF8
|
PHD finger protein 8 |
| chr14_-_37051798 | 0.06 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr17_+_67498295 | 0.06 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr2_+_210444748 | 0.06 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_+_211433275 | 0.06 |
ENST00000367005.4
|
RCOR3
|
REST corepressor 3 |
| chr1_+_24646263 | 0.06 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr13_+_93879085 | 0.06 |
ENST00000377047.4
|
GPC6
|
glypican 6 |
| chr16_-_18468926 | 0.06 |
ENST00000545114.1
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr12_-_18890940 | 0.06 |
ENST00000543242.1
ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1
|
phospholipase C, zeta 1 |
| chr18_-_2982869 | 0.06 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr2_+_168725458 | 0.06 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr8_+_107738343 | 0.06 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr6_-_134499037 | 0.06 |
ENST00000528577.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr4_-_186732241 | 0.06 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_33168391 | 0.06 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr2_-_21266816 | 0.06 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr16_-_75282088 | 0.06 |
ENST00000542031.2
|
BCAR1
|
breast cancer anti-estrogen resistance 1 |
| chr16_+_89724188 | 0.06 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr1_+_104293028 | 0.06 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr17_+_48823975 | 0.05 |
ENST00000513969.1
ENST00000503728.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr11_+_102980251 | 0.05 |
ENST00000334267.7
ENST00000398093.3 |
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr11_-_7950209 | 0.05 |
ENST00000309838.2
|
OR10A6
|
olfactory receptor, family 10, subfamily A, member 6 |
| chr5_+_149877440 | 0.05 |
ENST00000518299.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr10_-_29811456 | 0.05 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr8_-_114449112 | 0.05 |
ENST00000455883.2
ENST00000352409.3 ENST00000297405.5 |
CSMD3
|
CUB and Sushi multiple domains 3 |
| chrY_+_4868267 | 0.05 |
ENST00000333703.4
|
PCDH11Y
|
protocadherin 11 Y-linked |
| chr12_-_88423164 | 0.05 |
ENST00000298699.2
ENST00000550553.1 |
C12orf50
|
chromosome 12 open reading frame 50 |
| chr8_+_16884740 | 0.05 |
ENST00000318063.5
|
MICU3
|
mitochondrial calcium uptake family, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) positive regulation of mitochondrial DNA metabolic process(GO:1901860) response to methionine(GO:1904640) |
| 0.2 | 0.7 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) regulation of positive thymic T cell selection(GO:1902232) |
| 0.1 | 0.6 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 0.2 | GO:1902960 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.5 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.6 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.4 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.3 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.2 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.7 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.0 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.2 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.0 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.7 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |