Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CUX2
Z-value: 0.57
Transcription factors associated with CUX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX2
|
ENSG00000111249.9 | cut like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg19_v2_chr12_+_111471828_111471975 | -0.49 | 3.3e-01 | Click! |
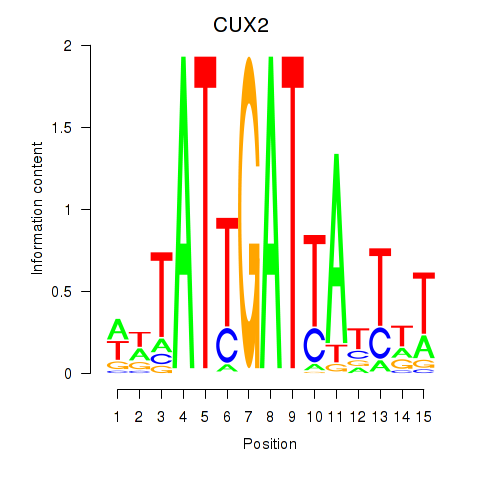
Activity profile of CUX2 motif
Sorted Z-values of CUX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_119669160 | 0.41 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr11_+_110225855 | 0.37 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr22_-_28392227 | 0.32 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr10_+_6779326 | 0.31 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr16_-_75467274 | 0.30 |
ENST00000566254.1
|
CFDP1
|
craniofacial development protein 1 |
| chr6_+_46761118 | 0.29 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr14_-_81425828 | 0.29 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr8_-_65730127 | 0.25 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr2_+_9778872 | 0.23 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr8_-_82395461 | 0.22 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr1_+_81106951 | 0.21 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr10_+_35484793 | 0.21 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr13_-_46742630 | 0.21 |
ENST00000416500.1
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_+_101659682 | 0.20 |
ENST00000465215.1
|
RP11-221J22.1
|
RP11-221J22.1 |
| chr19_+_46003056 | 0.19 |
ENST00000401593.1
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr2_-_214017151 | 0.19 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chrY_+_15418467 | 0.19 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr14_-_73997901 | 0.19 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr9_+_131683174 | 0.18 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr16_-_70239683 | 0.18 |
ENST00000601706.1
|
AC009060.1
|
Uncharacterized protein |
| chr5_-_54988559 | 0.18 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr20_-_45530365 | 0.17 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr11_-_104480019 | 0.17 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr6_-_15548591 | 0.17 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr19_-_58864848 | 0.17 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chrX_-_80457385 | 0.16 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr6_+_31105426 | 0.16 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr20_+_62492566 | 0.15 |
ENST00000369916.3
|
ABHD16B
|
abhydrolase domain containing 16B |
| chr19_+_29704142 | 0.15 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr4_-_69083720 | 0.15 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr18_+_52385068 | 0.14 |
ENST00000586570.1
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr14_+_50159813 | 0.14 |
ENST00000359332.2
ENST00000553274.1 ENST00000557128.1 |
KLHDC1
|
kelch domain containing 1 |
| chr13_-_46756351 | 0.14 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_-_19615744 | 0.14 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr9_+_131684027 | 0.14 |
ENST00000426694.1
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr5_-_135290705 | 0.14 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr22_+_30821732 | 0.13 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr1_+_149239529 | 0.13 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr8_-_38008783 | 0.13 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr3_-_100565249 | 0.13 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_-_4720301 | 0.13 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr21_+_39668478 | 0.12 |
ENST00000398927.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_105010320 | 0.12 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr8_+_22601 | 0.12 |
ENST00000522481.3
ENST00000518652.1 |
AC144568.2
|
Uncharacterized protein |
| chr12_+_42624050 | 0.12 |
ENST00000601185.1
|
AC020629.1
|
Uncharacterized protein |
| chrX_+_54947229 | 0.12 |
ENST00000442098.1
ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO
|
trophinin |
| chr13_+_21714653 | 0.12 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr12_+_123944070 | 0.11 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr1_-_151804314 | 0.11 |
ENST00000318247.6
|
RORC
|
RAR-related orphan receptor C |
| chr12_-_106477805 | 0.11 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr11_+_34999328 | 0.11 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr19_+_7011509 | 0.11 |
ENST00000377296.3
|
AC025278.1
|
Uncharacterized protein |
| chr11_+_134144139 | 0.11 |
ENST00000389887.5
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr7_-_33842742 | 0.11 |
ENST00000420185.1
ENST00000440034.1 |
RP11-89N17.4
|
RP11-89N17.4 |
| chr6_-_31613280 | 0.11 |
ENST00000453833.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr12_-_11422739 | 0.11 |
ENST00000279573.7
|
PRB3
|
proline-rich protein BstNI subfamily 3 |
| chr3_+_67705121 | 0.11 |
ENST00000464420.1
ENST00000482677.1 |
RP11-81N13.1
|
RP11-81N13.1 |
| chr2_+_47454054 | 0.11 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr20_-_22566089 | 0.11 |
ENST00000377115.4
|
FOXA2
|
forkhead box A2 |
| chr2_+_132233664 | 0.11 |
ENST00000321253.6
|
TUBA3D
|
tubulin, alpha 3d |
| chr3_+_172034218 | 0.10 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr19_-_22379753 | 0.10 |
ENST00000397121.2
|
ZNF676
|
zinc finger protein 676 |
| chr9_+_140445651 | 0.10 |
ENST00000371443.5
|
MRPL41
|
mitochondrial ribosomal protein L41 |
| chr20_+_31755934 | 0.10 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr1_-_9953295 | 0.10 |
ENST00000377258.1
|
CTNNBIP1
|
catenin, beta interacting protein 1 |
| chr1_-_23340400 | 0.10 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr2_-_99917639 | 0.10 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr4_-_186570679 | 0.10 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_143771275 | 0.09 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr6_+_26273144 | 0.09 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chrX_+_1734051 | 0.09 |
ENST00000381229.4
ENST00000381233.3 |
ASMT
|
acetylserotonin O-methyltransferase |
| chr11_+_101983176 | 0.09 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr9_+_93921855 | 0.09 |
ENST00000423719.2
ENST00000609752.1 |
LINC00484
|
long intergenic non-protein coding RNA 484 |
| chr6_-_138539627 | 0.09 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr12_+_47610315 | 0.09 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr3_-_122512619 | 0.09 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr10_+_53806501 | 0.09 |
ENST00000373975.2
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr6_-_27799305 | 0.09 |
ENST00000357549.2
|
HIST1H4K
|
histone cluster 1, H4k |
| chr1_+_44514040 | 0.09 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr16_-_66584059 | 0.08 |
ENST00000417693.3
ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2
|
thymidine kinase 2, mitochondrial |
| chr15_-_51397473 | 0.08 |
ENST00000327536.5
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr7_+_125078119 | 0.08 |
ENST00000458437.1
ENST00000415896.1 |
RP11-807H17.1
|
RP11-807H17.1 |
| chr3_-_112693865 | 0.08 |
ENST00000471858.1
ENST00000295863.4 ENST00000308611.3 |
CD200R1
|
CD200 receptor 1 |
| chr3_+_142842128 | 0.08 |
ENST00000483262.1
|
RP11-80H8.4
|
RP11-80H8.4 |
| chr5_+_59726565 | 0.08 |
ENST00000412930.2
|
FKSG52
|
FKSG52 |
| chr10_+_70980051 | 0.08 |
ENST00000354624.5
ENST00000395086.2 |
HKDC1
|
hexokinase domain containing 1 |
| chr3_+_159557637 | 0.08 |
ENST00000445224.2
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr3_+_181429704 | 0.08 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr12_+_6554021 | 0.08 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr12_-_10605929 | 0.08 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr3_+_121289551 | 0.08 |
ENST00000334384.3
|
ARGFX
|
arginine-fifty homeobox |
| chr2_-_172291273 | 0.08 |
ENST00000442778.1
ENST00000453846.1 |
METTL8
|
methyltransferase like 8 |
| chr17_+_41150290 | 0.08 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr21_-_28215332 | 0.08 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr15_-_49170219 | 0.08 |
ENST00000558220.1
ENST00000537958.1 |
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr19_-_52674896 | 0.08 |
ENST00000322146.8
ENST00000597065.1 |
ZNF836
|
zinc finger protein 836 |
| chr5_+_36606992 | 0.08 |
ENST00000505202.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_+_67498538 | 0.07 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr14_+_21566980 | 0.07 |
ENST00000418511.2
ENST00000554329.2 |
TMEM253
|
transmembrane protein 253 |
| chr1_+_8378140 | 0.07 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr4_-_144940477 | 0.07 |
ENST00000513128.1
ENST00000429670.2 ENST00000502664.1 |
GYPB
|
glycophorin B (MNS blood group) |
| chr19_+_48774586 | 0.07 |
ENST00000594024.1
ENST00000595408.1 ENST00000315849.1 |
ZNF114
|
zinc finger protein 114 |
| chrX_-_138072729 | 0.07 |
ENST00000455663.1
|
FGF13
|
fibroblast growth factor 13 |
| chr5_-_133706695 | 0.07 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr22_+_45148432 | 0.07 |
ENST00000389774.2
ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8
|
Rho GTPase activating protein 8 |
| chr11_+_122709200 | 0.07 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr17_-_34890759 | 0.07 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr2_+_102953608 | 0.07 |
ENST00000311734.2
ENST00000409584.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr3_+_186158169 | 0.07 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr5_+_119799927 | 0.07 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr10_-_90967063 | 0.07 |
ENST00000371852.2
|
CH25H
|
cholesterol 25-hydroxylase |
| chr12_-_7596735 | 0.07 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr15_+_89631647 | 0.07 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr17_-_34890732 | 0.07 |
ENST00000268852.9
|
MYO19
|
myosin XIX |
| chrX_-_73051037 | 0.07 |
ENST00000445814.1
|
XIST
|
X inactive specific transcript (non-protein coding) |
| chr1_+_87458692 | 0.07 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chrM_+_4431 | 0.07 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_+_49518113 | 0.07 |
ENST00000529246.2
|
C6orf141
|
chromosome 6 open reading frame 141 |
| chr1_-_109935819 | 0.07 |
ENST00000538502.1
|
SORT1
|
sortilin 1 |
| chr10_-_419129 | 0.07 |
ENST00000540204.1
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr16_-_28634874 | 0.07 |
ENST00000395609.1
ENST00000350842.4 |
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr8_+_95558771 | 0.07 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr3_-_28389922 | 0.07 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr7_+_102290772 | 0.07 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr19_-_37697976 | 0.06 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
| chr3_+_157827841 | 0.06 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_108321623 | 0.06 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr21_+_39644305 | 0.06 |
ENST00000398930.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr8_+_93895865 | 0.06 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr4_-_105416039 | 0.06 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr5_-_1801408 | 0.06 |
ENST00000505818.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr2_+_234600253 | 0.06 |
ENST00000373424.1
ENST00000441351.1 |
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr16_-_66583994 | 0.06 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr4_-_85654615 | 0.06 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr10_-_127505167 | 0.06 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr4_-_70518941 | 0.06 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr3_+_157828152 | 0.06 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chrX_-_117119243 | 0.06 |
ENST00000539496.1
ENST00000469946.1 |
KLHL13
|
kelch-like family member 13 |
| chr17_-_3595181 | 0.06 |
ENST00000552050.1
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr21_+_39644214 | 0.06 |
ENST00000438657.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_82745238 | 0.06 |
ENST00000531021.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr5_-_57854070 | 0.06 |
ENST00000504333.1
|
CTD-2117L12.1
|
Uncharacterized protein |
| chr6_+_28249299 | 0.06 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr21_-_43346790 | 0.06 |
ENST00000329623.7
|
C2CD2
|
C2 calcium-dependent domain containing 2 |
| chrX_-_63615297 | 0.06 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chrX_-_30595959 | 0.06 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr2_+_217082311 | 0.06 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chr6_-_133084580 | 0.06 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr16_-_30621663 | 0.06 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr2_+_27886330 | 0.06 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr11_+_44117219 | 0.06 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr11_+_58938903 | 0.06 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr9_+_74729511 | 0.06 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr10_-_90751038 | 0.06 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr5_+_140501581 | 0.06 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr10_-_28270795 | 0.06 |
ENST00000545014.1
|
ARMC4
|
armadillo repeat containing 4 |
| chrX_+_55246818 | 0.06 |
ENST00000374952.1
|
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr19_+_44645700 | 0.06 |
ENST00000592437.1
|
ZNF234
|
zinc finger protein 234 |
| chr1_-_153950098 | 0.06 |
ENST00000356648.1
|
JTB
|
jumping translocation breakpoint |
| chr1_+_186265399 | 0.06 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr19_-_39402798 | 0.06 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr4_+_147145709 | 0.06 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr20_-_58515344 | 0.06 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr2_-_220264703 | 0.06 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr4_-_17513604 | 0.06 |
ENST00000505710.1
|
QDPR
|
quinoid dihydropteridine reductase |
| chr16_-_75467318 | 0.05 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr3_-_48598547 | 0.05 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr19_-_55677999 | 0.05 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr3_+_169539710 | 0.05 |
ENST00000340806.6
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4 |
| chr11_+_5775923 | 0.05 |
ENST00000317254.3
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4 (gene/pseudogene) |
| chr10_+_90484301 | 0.05 |
ENST00000404190.1
|
LIPK
|
lipase, family member K |
| chr22_+_38071615 | 0.05 |
ENST00000215909.5
|
LGALS1
|
lectin, galactoside-binding, soluble, 1 |
| chr14_+_21458127 | 0.05 |
ENST00000382985.4
ENST00000556670.2 ENST00000553564.1 ENST00000554751.1 ENST00000554283.1 ENST00000555670.1 |
METTL17
|
methyltransferase like 17 |
| chr5_+_60933634 | 0.05 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr19_-_29704448 | 0.05 |
ENST00000304863.4
|
UQCRFS1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr1_+_152850787 | 0.05 |
ENST00000368765.3
|
SMCP
|
sperm mitochondria-associated cysteine-rich protein |
| chr17_-_4167142 | 0.05 |
ENST00000570535.1
ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1
|
ankyrin repeat and FYVE domain containing 1 |
| chr22_+_18560743 | 0.05 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr5_+_128300810 | 0.05 |
ENST00000262462.4
|
SLC27A6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr1_+_43291220 | 0.05 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr2_+_181988620 | 0.05 |
ENST00000428474.1
ENST00000424655.1 |
AC104820.2
|
AC104820.2 |
| chr5_+_173472607 | 0.05 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr1_-_54304212 | 0.05 |
ENST00000540001.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chrX_+_8432871 | 0.05 |
ENST00000381032.1
ENST00000453306.1 ENST00000444481.1 |
VCX3B
|
variable charge, X-linked 3B |
| chr19_+_58193388 | 0.05 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr1_-_153950116 | 0.05 |
ENST00000368589.1
|
JTB
|
jumping translocation breakpoint |
| chr5_-_43397184 | 0.05 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr17_-_18430160 | 0.05 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr9_-_138853156 | 0.05 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr6_-_30658745 | 0.05 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr12_+_74931551 | 0.05 |
ENST00000519948.2
|
ATXN7L3B
|
ataxin 7-like 3B |
| chr16_+_58283814 | 0.05 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr13_+_21714711 | 0.05 |
ENST00000607003.1
ENST00000492245.1 |
SAP18
|
Sin3A-associated protein, 18kDa |
| chr3_-_197300194 | 0.05 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr5_+_121465207 | 0.05 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr12_+_43086018 | 0.05 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr8_-_8318847 | 0.05 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr17_+_67498396 | 0.05 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chrX_+_55246771 | 0.05 |
ENST00000289619.5
ENST00000374955.3 |
PAGE5
|
P antigen family, member 5 (prostate associated) |
| chr4_-_100575781 | 0.05 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018963 | insecticide metabolic process(GO:0017143) phthalate metabolic process(GO:0018963) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.4 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0046092 | deoxycytidine metabolic process(GO:0046092) |
| 0.0 | 0.1 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0070408 | carbamoyl phosphate metabolic process(GO:0070408) carbamoyl phosphate biosynthetic process(GO:0070409) response to ammonia(GO:1903717) cellular response to ammonia(GO:1903718) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.0 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.0 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.0 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.0 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.0 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.0 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |