Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
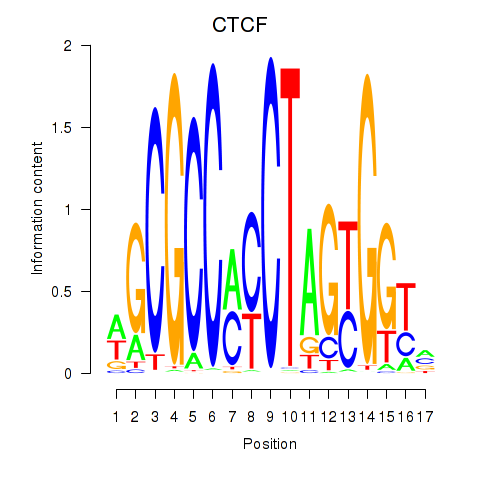
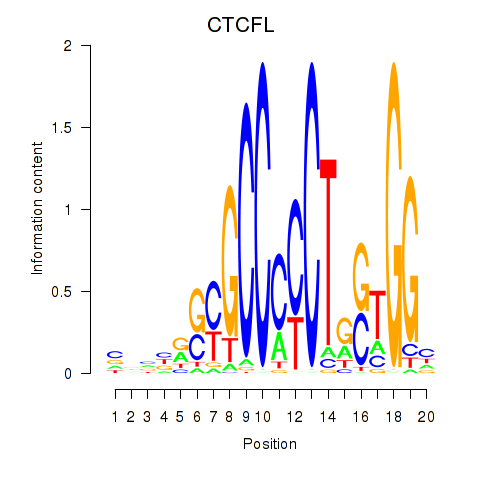
Results for CTCF_CTCFL
Z-value: 0.57
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CTCF
|
ENSG00000102974.10 | CCCTC-binding factor |
|
CTCFL
|
ENSG00000124092.8 | CCCTC-binding factor like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCF | hg19_v2_chr16_+_67596310_67596353 | -0.40 | 4.3e-01 | Click! |
| CTCFL | hg19_v2_chr20_-_56100155_56100163 | -0.26 | 6.1e-01 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_114737146 | 0.86 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chr2_-_232571621 | 0.61 |
ENST00000595658.1
|
MGC4771
|
MGC4771 |
| chr7_-_100171270 | 0.59 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr2_+_130939235 | 0.54 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr2_-_132249955 | 0.53 |
ENST00000309451.6
|
MZT2A
|
mitotic spindle organizing protein 2A |
| chr15_-_64338521 | 0.45 |
ENST00000457488.1
ENST00000558069.1 |
DAPK2
|
death-associated protein kinase 2 |
| chr17_+_43213004 | 0.42 |
ENST00000586346.1
ENST00000398322.3 ENST00000592162.1 ENST00000376955.4 ENST00000321854.8 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr8_-_48651648 | 0.42 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr10_+_99332198 | 0.37 |
ENST00000307518.5
ENST00000298808.5 ENST00000370655.1 |
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr19_+_5823813 | 0.36 |
ENST00000303212.2
|
NRTN
|
neurturin |
| chr9_+_140317802 | 0.36 |
ENST00000341349.2
ENST00000392815.2 |
NOXA1
|
NADPH oxidase activator 1 |
| chr2_+_130939827 | 0.32 |
ENST00000409255.1
ENST00000455239.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr20_+_36531499 | 0.30 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr22_-_50765489 | 0.29 |
ENST00000413817.3
|
DENND6B
|
DENN/MADD domain containing 6B |
| chr12_-_8549301 | 0.28 |
ENST00000544461.1
|
LINC00937
|
long intergenic non-protein coding RNA 937 |
| chr4_+_89378261 | 0.28 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr9_-_22009241 | 0.28 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr19_-_4400415 | 0.26 |
ENST00000598564.1
ENST00000417295.2 ENST00000269886.3 |
SH3GL1
|
SH3-domain GRB2-like 1 |
| chr19_+_507299 | 0.26 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr16_+_1756162 | 0.25 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr2_+_114737472 | 0.24 |
ENST00000420161.1
|
AC110769.3
|
AC110769.3 |
| chr19_-_1848451 | 0.24 |
ENST00000170168.4
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr9_+_127539425 | 0.23 |
ENST00000331715.9
|
OLFML2A
|
olfactomedin-like 2A |
| chr20_+_49411523 | 0.23 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr9_+_127539481 | 0.23 |
ENST00000373580.3
|
OLFML2A
|
olfactomedin-like 2A |
| chr20_-_33460621 | 0.23 |
ENST00000427420.1
ENST00000336431.5 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr5_+_116790848 | 0.22 |
ENST00000504107.1
|
LINC00992
|
long intergenic non-protein coding RNA 992 |
| chr7_-_16840820 | 0.22 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr19_+_38893809 | 0.20 |
ENST00000589408.1
|
FAM98C
|
family with sequence similarity 98, member C |
| chr16_+_48657337 | 0.20 |
ENST00000568150.1
ENST00000564212.1 |
RP11-42I10.1
|
RP11-42I10.1 |
| chr9_-_140317605 | 0.20 |
ENST00000479452.1
ENST00000465160.2 |
EXD3
|
exonuclease 3'-5' domain containing 3 |
| chr11_-_62379752 | 0.20 |
ENST00000466671.1
ENST00000466886.1 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr9_-_139940608 | 0.19 |
ENST00000371601.4
|
NPDC1
|
neural proliferation, differentiation and control, 1 |
| chr9_+_92219919 | 0.19 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr1_-_159915386 | 0.19 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr15_+_76352178 | 0.19 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr22_+_24204375 | 0.19 |
ENST00000433835.3
|
AP000350.10
|
Uncharacterized protein |
| chr19_+_7745708 | 0.19 |
ENST00000596148.1
ENST00000317378.5 ENST00000426877.2 |
TRAPPC5
|
trafficking protein particle complex 5 |
| chr19_-_58400372 | 0.18 |
ENST00000597832.1
ENST00000435989.2 |
ZNF814
|
zinc finger protein 814 |
| chr9_-_139922726 | 0.18 |
ENST00000265662.5
ENST00000371605.3 |
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr17_+_78075361 | 0.18 |
ENST00000577106.1
ENST00000390015.3 |
GAA
|
glucosidase, alpha; acid |
| chr4_+_25314388 | 0.17 |
ENST00000302874.4
|
ZCCHC4
|
zinc finger, CCHC domain containing 4 |
| chr16_+_67313412 | 0.17 |
ENST00000379344.3
ENST00000568621.1 ENST00000450733.1 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr2_+_136499287 | 0.17 |
ENST00000415164.1
|
UBXN4
|
UBX domain protein 4 |
| chr11_-_117102768 | 0.17 |
ENST00000532301.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr20_+_43992094 | 0.17 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr9_+_131464767 | 0.17 |
ENST00000291906.4
|
PKN3
|
protein kinase N3 |
| chr9_-_139922631 | 0.16 |
ENST00000341511.6
|
ABCA2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr15_-_65809625 | 0.16 |
ENST00000560436.1
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr10_+_99332529 | 0.16 |
ENST00000455090.1
|
ANKRD2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr6_+_3849584 | 0.16 |
ENST00000380274.1
ENST00000380272.3 |
FAM50B
|
family with sequence similarity 50, member B |
| chr1_-_1284730 | 0.16 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr17_+_78075498 | 0.16 |
ENST00000302262.3
|
GAA
|
glucosidase, alpha; acid |
| chr3_+_48507210 | 0.16 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr12_+_48099858 | 0.16 |
ENST00000547799.1
|
RP1-197B17.3
|
RP1-197B17.3 |
| chr16_+_30996502 | 0.15 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr20_+_44462749 | 0.15 |
ENST00000372541.1
|
SNX21
|
sorting nexin family member 21 |
| chr17_+_63096903 | 0.15 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr19_-_48823332 | 0.15 |
ENST00000315396.7
|
CCDC114
|
coiled-coil domain containing 114 |
| chr17_-_61523622 | 0.15 |
ENST00000448884.2
ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561
|
cytochrome b561 |
| chr3_+_49209023 | 0.15 |
ENST00000332780.2
|
KLHDC8B
|
kelch domain containing 8B |
| chr11_+_46368956 | 0.14 |
ENST00000543978.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr14_+_101293687 | 0.14 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr5_+_176513895 | 0.14 |
ENST00000503708.1
ENST00000393648.2 ENST00000514472.1 ENST00000502906.1 ENST00000292410.3 ENST00000510911.1 |
FGFR4
|
fibroblast growth factor receptor 4 |
| chr19_-_44008863 | 0.14 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr11_-_66103932 | 0.14 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr17_+_16593539 | 0.14 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr8_-_145911188 | 0.14 |
ENST00000540274.1
|
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr2_+_220306238 | 0.14 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr9_-_117160738 | 0.14 |
ENST00000448674.1
|
RP11-9M16.2
|
RP11-9M16.2 |
| chr11_-_65359947 | 0.14 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chrX_+_77166172 | 0.13 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr9_+_19408919 | 0.13 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr20_+_44462430 | 0.13 |
ENST00000462307.1
|
SNX21
|
sorting nexin family member 21 |
| chr16_-_3086927 | 0.13 |
ENST00000572449.1
|
CCDC64B
|
coiled-coil domain containing 64B |
| chr1_-_40367668 | 0.13 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_-_66104237 | 0.13 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr19_-_59031118 | 0.13 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr16_+_230435 | 0.13 |
ENST00000199708.2
|
HBQ1
|
hemoglobin, theta 1 |
| chr19_-_50990785 | 0.13 |
ENST00000595005.1
|
CTD-2545M3.8
|
CTD-2545M3.8 |
| chr20_-_34542548 | 0.12 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr19_+_49977466 | 0.12 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr5_+_140254884 | 0.12 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chrX_+_153029633 | 0.12 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr22_-_19132154 | 0.12 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr9_-_130497565 | 0.12 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr15_-_75748143 | 0.12 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr1_-_40367530 | 0.12 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_-_128775930 | 0.11 |
ENST00000524878.1
|
C11orf45
|
chromosome 11 open reading frame 45 |
| chr11_+_61447845 | 0.11 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr5_-_159739483 | 0.11 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr4_+_81256871 | 0.11 |
ENST00000358105.3
ENST00000508675.1 |
C4orf22
|
chromosome 4 open reading frame 22 |
| chr17_+_74864476 | 0.11 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr17_+_73629500 | 0.11 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr22_+_24551765 | 0.11 |
ENST00000337989.7
|
CABIN1
|
calcineurin binding protein 1 |
| chr5_+_176513868 | 0.11 |
ENST00000292408.4
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr11_-_59633951 | 0.11 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr2_+_181845532 | 0.11 |
ENST00000602475.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr19_-_58666399 | 0.11 |
ENST00000601887.1
|
ZNF329
|
zinc finger protein 329 |
| chr17_-_73511584 | 0.11 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr8_+_22446763 | 0.11 |
ENST00000450780.2
ENST00000430850.2 ENST00000447849.1 |
AC037459.4
|
Uncharacterized protein |
| chr19_-_45996465 | 0.11 |
ENST00000430715.2
|
RTN2
|
reticulon 2 |
| chr19_+_42829702 | 0.11 |
ENST00000334370.4
|
MEGF8
|
multiple EGF-like-domains 8 |
| chr17_-_77813186 | 0.10 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr3_+_184279566 | 0.10 |
ENST00000330394.2
|
EPHB3
|
EPH receptor B3 |
| chr20_+_49411543 | 0.10 |
ENST00000609336.1
ENST00000445038.1 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr15_+_75074385 | 0.10 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr16_+_4674814 | 0.10 |
ENST00000415496.1
ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr11_+_119038897 | 0.10 |
ENST00000454811.1
ENST00000449394.1 |
NLRX1
|
NLR family member X1 |
| chr12_+_13197218 | 0.10 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr17_-_33446735 | 0.10 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr19_+_5690207 | 0.10 |
ENST00000347512.3
|
RPL36
|
ribosomal protein L36 |
| chr9_-_134615326 | 0.10 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chrX_-_99891796 | 0.10 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr15_+_71185148 | 0.10 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr6_+_139349903 | 0.10 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr17_+_78075324 | 0.10 |
ENST00000570803.1
|
GAA
|
glucosidase, alpha; acid |
| chr19_-_49339080 | 0.10 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr20_+_44462481 | 0.10 |
ENST00000491381.1
ENST00000342644.5 ENST00000372542.1 |
SNX21
|
sorting nexin family member 21 |
| chr19_+_5690297 | 0.10 |
ENST00000582463.1
ENST00000579446.1 ENST00000394580.2 |
RPL36
|
ribosomal protein L36 |
| chr9_-_35096545 | 0.10 |
ENST00000378617.3
ENST00000341666.3 ENST00000361778.2 |
PIGO
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr14_+_23341513 | 0.10 |
ENST00000546834.1
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr3_-_186857267 | 0.10 |
ENST00000455270.1
ENST00000296277.4 |
RPL39L
|
ribosomal protein L39-like |
| chr14_+_54976546 | 0.09 |
ENST00000216420.7
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr11_+_60691924 | 0.09 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr14_-_102701740 | 0.09 |
ENST00000561150.1
ENST00000522867.1 |
MOK
|
MOK protein kinase |
| chr20_-_30539773 | 0.09 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr17_-_73511504 | 0.09 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr3_-_49203744 | 0.09 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr11_-_8285405 | 0.09 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr11_+_67033881 | 0.09 |
ENST00000308595.5
ENST00000526285.1 |
ADRBK1
|
adrenergic, beta, receptor kinase 1 |
| chr9_-_130742792 | 0.09 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr2_+_232575168 | 0.09 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr6_-_32920794 | 0.09 |
ENST00000395305.3
ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA
XXbac-BPG181M17.5
|
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr16_+_78133536 | 0.09 |
ENST00000402655.2
ENST00000406884.2 ENST00000539474.2 ENST00000569818.1 ENST00000355860.3 ENST00000408984.3 |
WWOX
|
WW domain containing oxidoreductase |
| chr9_+_116638630 | 0.09 |
ENST00000452710.1
ENST00000374124.4 |
ZNF618
|
zinc finger protein 618 |
| chr8_-_145550571 | 0.09 |
ENST00000332324.4
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr5_-_76788298 | 0.09 |
ENST00000511036.1
|
WDR41
|
WD repeat domain 41 |
| chr5_+_140514782 | 0.09 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr17_+_47653178 | 0.09 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr6_-_44225231 | 0.09 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr19_-_59030921 | 0.09 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr16_-_80838160 | 0.09 |
ENST00000562812.1
ENST00000563890.1 ENST00000566173.1 |
CDYL2
|
chromodomain protein, Y-like 2 |
| chr15_-_65810042 | 0.09 |
ENST00000321147.6
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr14_+_105941118 | 0.09 |
ENST00000550577.1
ENST00000538259.2 |
CRIP2
|
cysteine-rich protein 2 |
| chr9_+_100263912 | 0.09 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr16_+_4674787 | 0.09 |
ENST00000262370.7
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr14_-_36990354 | 0.09 |
ENST00000518149.1
|
NKX2-1
|
NK2 homeobox 1 |
| chr22_-_31503490 | 0.08 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr14_-_91526462 | 0.08 |
ENST00000536315.2
|
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr19_+_18043810 | 0.08 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr13_+_96204961 | 0.08 |
ENST00000299339.2
|
CLDN10
|
claudin 10 |
| chr9_+_100174344 | 0.08 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr11_+_2421718 | 0.08 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr14_+_55738021 | 0.08 |
ENST00000313833.4
|
FBXO34
|
F-box protein 34 |
| chr4_+_980785 | 0.08 |
ENST00000247933.4
ENST00000453894.1 |
IDUA
|
iduronidase, alpha-L- |
| chr17_+_7591747 | 0.08 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr18_+_60382672 | 0.08 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr10_-_71906342 | 0.08 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr4_-_2010562 | 0.08 |
ENST00000411649.1
ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA
|
negative elongation factor complex member A |
| chr9_+_131182697 | 0.08 |
ENST00000372838.4
ENST00000411852.1 |
CERCAM
|
cerebral endothelial cell adhesion molecule |
| chr16_+_71392616 | 0.08 |
ENST00000349553.5
ENST00000302628.4 ENST00000562305.1 |
CALB2
|
calbindin 2 |
| chr1_+_10490779 | 0.08 |
ENST00000477755.1
|
APITD1
|
apoptosis-inducing, TAF9-like domain 1 |
| chr3_-_183543301 | 0.08 |
ENST00000318631.3
ENST00000431348.1 |
MAP6D1
|
MAP6 domain containing 1 |
| chr19_-_54676846 | 0.08 |
ENST00000301187.4
|
TMC4
|
transmembrane channel-like 4 |
| chr7_-_91509986 | 0.08 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr11_+_58938903 | 0.08 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr11_+_73675873 | 0.08 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr19_-_4065730 | 0.08 |
ENST00000601588.1
|
ZBTB7A
|
zinc finger and BTB domain containing 7A |
| chr7_+_148844516 | 0.08 |
ENST00000420008.2
ENST00000475153.1 |
ZNF398
|
zinc finger protein 398 |
| chr16_+_67233412 | 0.08 |
ENST00000477898.1
|
ELMO3
|
engulfment and cell motility 3 |
| chr10_-_82116497 | 0.08 |
ENST00000372204.3
|
DYDC1
|
DPY30 domain containing 1 |
| chr11_-_72504637 | 0.08 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr11_-_798305 | 0.08 |
ENST00000531514.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr12_+_65996599 | 0.07 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr15_+_75074410 | 0.07 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr20_-_25038804 | 0.07 |
ENST00000323482.4
|
ACSS1
|
acyl-CoA synthetase short-chain family member 1 |
| chr5_+_140186647 | 0.07 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr19_-_14682838 | 0.07 |
ENST00000215565.2
|
NDUFB7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa |
| chr1_+_10490441 | 0.07 |
ENST00000470413.2
ENST00000309048.3 |
APITD1-CORT
APITD1
|
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr16_+_29827285 | 0.07 |
ENST00000320330.6
|
PAGR1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr4_+_154265784 | 0.07 |
ENST00000240488.3
|
MND1
|
meiotic nuclear divisions 1 homolog (S. cerevisiae) |
| chr19_+_48216600 | 0.07 |
ENST00000263277.3
ENST00000538399.1 |
EHD2
|
EH-domain containing 2 |
| chr19_-_54676884 | 0.07 |
ENST00000376591.4
|
TMC4
|
transmembrane channel-like 4 |
| chr1_+_154378049 | 0.07 |
ENST00000512471.1
|
IL6R
|
interleukin 6 receptor |
| chr2_-_86116020 | 0.07 |
ENST00000525834.2
|
ST3GAL5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr10_-_15762124 | 0.07 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr17_+_17584763 | 0.07 |
ENST00000353383.1
|
RAI1
|
retinoic acid induced 1 |
| chr15_-_65809991 | 0.07 |
ENST00000559526.1
ENST00000358939.4 ENST00000560665.1 ENST00000321118.7 ENST00000339244.5 ENST00000300141.6 |
DPP8
|
dipeptidyl-peptidase 8 |
| chr17_+_73996987 | 0.07 |
ENST00000588812.1
ENST00000448471.1 |
CDK3
|
cyclin-dependent kinase 3 |
| chr3_+_184081137 | 0.07 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_+_52007693 | 0.07 |
ENST00000494478.1
|
ABHD14A
|
abhydrolase domain containing 14A |
| chr17_-_42441204 | 0.07 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr4_-_141348789 | 0.07 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr6_-_149867122 | 0.07 |
ENST00000253329.2
|
PPIL4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr18_+_77623668 | 0.07 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr4_+_4861385 | 0.07 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr4_-_104020968 | 0.07 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr3_-_183735651 | 0.07 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr19_-_38747172 | 0.07 |
ENST00000347262.4
ENST00000591585.1 ENST00000301242.4 |
PPP1R14A
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 0.5 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.3 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.6 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.0 | 0.1 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0042414 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.3 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.3 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.3 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.0 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.0 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) L-kynurenine metabolic process(GO:0097052) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.0 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0019427 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:0033686 | extraocular skeletal muscle development(GO:0002074) positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.3 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0047536 | 2-aminoadipate transaminase activity(GO:0047536) |
| 0.0 | 0.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.1 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.2 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.0 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.0 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |