Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
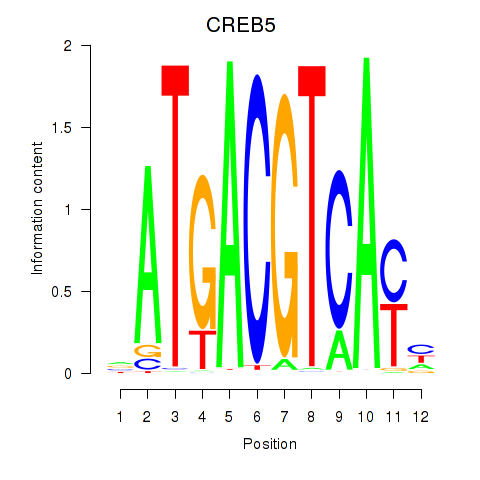
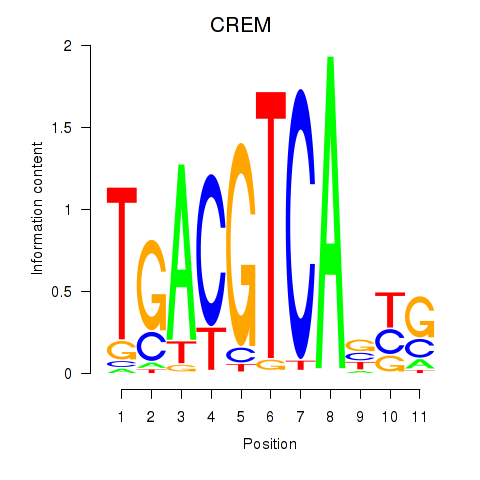
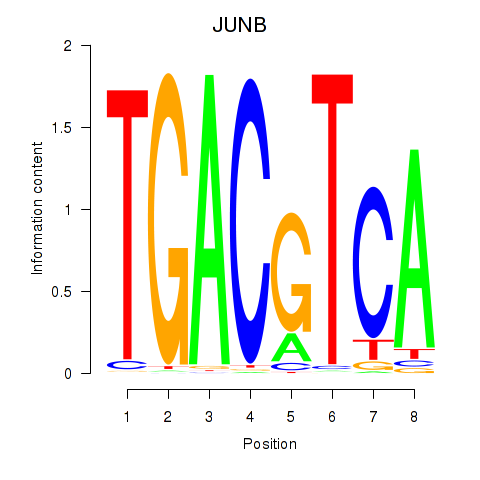
Results for CREB5_CREM_JUNB
Z-value: 0.66
Transcription factors associated with CREB5_CREM_JUNB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB5
|
ENSG00000146592.12 | cAMP responsive element binding protein 5 |
|
CREM
|
ENSG00000095794.15 | cAMP responsive element modulator |
|
JUNB
|
ENSG00000171223.4 | JunB proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREM | hg19_v2_chr10_+_35415851_35415941 | 0.86 | 2.7e-02 | Click! |
| CREB5 | hg19_v2_chr7_+_28448995_28449011 | 0.62 | 1.9e-01 | Click! |
| JUNB | hg19_v2_chr19_+_12902289_12902310 | -0.60 | 2.1e-01 | Click! |
Activity profile of CREB5_CREM_JUNB motif
Sorted Z-values of CREB5_CREM_JUNB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26204825 | 0.54 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr9_-_99381660 | 0.49 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr7_+_30174668 | 0.47 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr15_+_75287861 | 0.40 |
ENST00000425597.3
ENST00000562327.1 ENST00000568018.1 ENST00000562212.1 ENST00000567920.1 ENST00000566872.1 ENST00000361900.6 ENST00000545456.1 |
SCAMP5
|
secretory carrier membrane protein 5 |
| chr2_+_220408724 | 0.33 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr20_+_55099542 | 0.32 |
ENST00000371328.3
|
FAM209A
|
family with sequence similarity 209, member A |
| chr14_+_68086515 | 0.31 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr1_-_203274387 | 0.31 |
ENST00000432511.1
|
RP11-134P9.1
|
long intergenic non-protein coding RNA 1136 |
| chr11_+_64004888 | 0.30 |
ENST00000541681.1
|
VEGFB
|
vascular endothelial growth factor B |
| chr19_+_1954632 | 0.30 |
ENST00000589350.1
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr7_+_30174426 | 0.29 |
ENST00000324453.8
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr7_+_30174574 | 0.29 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr3_-_101395936 | 0.28 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr17_-_43210580 | 0.28 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr9_-_99382065 | 0.27 |
ENST00000265659.2
ENST00000375241.1 ENST00000375236.1 |
CDC14B
|
cell division cycle 14B |
| chr22_+_31518938 | 0.26 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr3_+_159570722 | 0.25 |
ENST00000482804.1
|
SCHIP1
|
schwannomin interacting protein 1 |
| chr12_-_4754356 | 0.25 |
ENST00000540967.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr10_-_14920599 | 0.25 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr5_-_99870890 | 0.23 |
ENST00000499025.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr6_-_31774714 | 0.22 |
ENST00000375661.5
|
LSM2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_-_6388389 | 0.22 |
ENST00000578372.1
|
FAM220A
|
family with sequence similarity 220, member A |
| chr5_+_133707252 | 0.22 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr12_+_124997766 | 0.21 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr16_+_29911864 | 0.21 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr1_+_70820451 | 0.21 |
ENST00000361764.4
ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3
|
HERV-H LTR-associating 3 |
| chr17_-_7155775 | 0.20 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr20_+_5892147 | 0.20 |
ENST00000455042.1
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr6_+_28048753 | 0.20 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr7_+_28448995 | 0.19 |
ENST00000424599.1
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr20_+_33146510 | 0.19 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr15_-_66790146 | 0.19 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr15_+_34394257 | 0.18 |
ENST00000397766.2
|
PGBD4
|
piggyBac transposable element derived 4 |
| chr2_-_202645612 | 0.18 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr4_-_156297919 | 0.18 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr3_+_38206975 | 0.18 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr12_-_123380610 | 0.18 |
ENST00000535765.1
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr22_+_38597889 | 0.17 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr14_+_38065052 | 0.17 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr1_+_27719148 | 0.16 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr2_-_224467002 | 0.16 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr3_+_63898275 | 0.16 |
ENST00000538065.1
|
ATXN7
|
ataxin 7 |
| chr2_-_25194476 | 0.16 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr18_+_76829258 | 0.15 |
ENST00000588600.1
|
ATP9B
|
ATPase, class II, type 9B |
| chr7_+_102292796 | 0.15 |
ENST00000436228.2
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr8_+_26240666 | 0.15 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr12_-_4754339 | 0.15 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr5_+_32174483 | 0.15 |
ENST00000606994.1
|
CTD-2186M15.3
|
CTD-2186M15.3 |
| chr6_-_37225391 | 0.14 |
ENST00000356757.2
|
TMEM217
|
transmembrane protein 217 |
| chr21_+_39628780 | 0.14 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr7_+_120590803 | 0.14 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr3_-_130745403 | 0.14 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr12_-_112856623 | 0.13 |
ENST00000551291.2
|
RPL6
|
ribosomal protein L6 |
| chr22_+_32340481 | 0.13 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr3_-_9811674 | 0.13 |
ENST00000411972.1
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr7_-_37956409 | 0.13 |
ENST00000436072.2
|
SFRP4
|
secreted frizzled-related protein 4 |
| chr5_-_130868688 | 0.13 |
ENST00000504575.1
ENST00000513227.1 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr12_-_100660833 | 0.13 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr19_+_10527449 | 0.13 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr1_-_205391178 | 0.13 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr16_-_11367452 | 0.13 |
ENST00000327157.2
|
PRM3
|
protamine 3 |
| chr11_+_66059339 | 0.12 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr3_+_170075436 | 0.12 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr8_-_116681686 | 0.12 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_+_18720316 | 0.12 |
ENST00000280734.2
|
TMEM86A
|
transmembrane protein 86A |
| chr17_+_33914460 | 0.12 |
ENST00000537622.2
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr22_+_32340447 | 0.12 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr1_+_85527987 | 0.12 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chrX_+_152224766 | 0.12 |
ENST00000370265.4
ENST00000447306.1 |
PNMA3
|
paraneoplastic Ma antigen 3 |
| chr6_-_121655850 | 0.12 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr9_+_70856899 | 0.12 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr4_+_1873100 | 0.11 |
ENST00000508803.1
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr20_+_44420570 | 0.11 |
ENST00000372622.3
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr5_-_78809950 | 0.11 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chrX_+_102024075 | 0.11 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr8_-_18711866 | 0.11 |
ENST00000519851.1
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr17_-_7835228 | 0.11 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr17_-_76899275 | 0.11 |
ENST00000322630.2
ENST00000586713.1 |
DDC8
|
Protein DDC8 homolog |
| chr11_-_62457371 | 0.11 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr3_-_156877997 | 0.11 |
ENST00000295926.3
|
CCNL1
|
cyclin L1 |
| chr17_+_29421900 | 0.11 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr14_-_93651186 | 0.10 |
ENST00000556883.1
ENST00000298894.4 |
MOAP1
|
modulator of apoptosis 1 |
| chr1_+_39456895 | 0.10 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr5_-_141703713 | 0.10 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr1_+_22379179 | 0.10 |
ENST00000315554.8
ENST00000421089.2 |
CDC42
|
cell division cycle 42 |
| chr2_-_224467093 | 0.10 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr20_-_44420507 | 0.10 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr2_+_11864458 | 0.10 |
ENST00000396098.1
ENST00000396099.1 ENST00000425416.2 |
LPIN1
|
lipin 1 |
| chr12_+_100660909 | 0.10 |
ENST00000549687.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr9_-_111696224 | 0.10 |
ENST00000537196.1
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr17_+_33914276 | 0.10 |
ENST00000592545.1
ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr7_-_140624499 | 0.10 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr17_-_40540377 | 0.10 |
ENST00000404395.3
ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr4_+_75311019 | 0.10 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr20_+_33292507 | 0.10 |
ENST00000414082.1
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr1_+_166808692 | 0.10 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr1_-_245026388 | 0.09 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr8_+_38243967 | 0.09 |
ENST00000524874.1
ENST00000379957.4 ENST00000523983.2 |
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr1_+_17531614 | 0.09 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chr7_+_120591170 | 0.09 |
ENST00000431467.1
|
ING3
|
inhibitor of growth family, member 3 |
| chr20_+_44420617 | 0.09 |
ENST00000449078.1
ENST00000456939.1 |
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chrX_-_152245978 | 0.09 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr19_-_50528584 | 0.09 |
ENST00000594092.1
ENST00000443401.2 ENST00000594948.1 ENST00000377011.2 ENST00000593919.1 ENST00000601324.1 ENST00000316763.3 ENST00000601341.1 ENST00000600259.1 |
VRK3
|
vaccinia related kinase 3 |
| chr4_+_668348 | 0.09 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr19_-_52227221 | 0.09 |
ENST00000222115.1
ENST00000540069.2 |
HAS1
|
hyaluronan synthase 1 |
| chr9_-_179018 | 0.09 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr22_-_39096925 | 0.09 |
ENST00000456626.1
ENST00000412832.1 |
JOSD1
|
Josephin domain containing 1 |
| chr11_+_3819049 | 0.09 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr5_-_39462390 | 0.09 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_-_25164969 | 0.09 |
ENST00000305786.2
|
CYCS
|
cytochrome c, somatic |
| chrX_-_135056106 | 0.09 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr16_+_22825475 | 0.09 |
ENST00000261374.3
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr19_-_50528392 | 0.09 |
ENST00000600137.1
ENST00000597215.1 |
VRK3
|
vaccinia related kinase 3 |
| chr19_+_16187085 | 0.09 |
ENST00000300933.4
|
TPM4
|
tropomyosin 4 |
| chr1_+_1115056 | 0.09 |
ENST00000379288.3
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10 |
| chr11_-_72496976 | 0.09 |
ENST00000539138.1
ENST00000542989.1 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr3_+_3168692 | 0.09 |
ENST00000402675.1
ENST00000413000.1 |
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1 |
| chr8_+_42128861 | 0.09 |
ENST00000518983.1
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr16_+_19079215 | 0.09 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr7_+_116660246 | 0.09 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr17_-_7165662 | 0.09 |
ENST00000571881.2
ENST00000360325.7 |
CLDN7
|
claudin 7 |
| chr1_-_209975494 | 0.09 |
ENST00000456314.1
|
IRF6
|
interferon regulatory factor 6 |
| chr7_-_25164868 | 0.09 |
ENST00000409409.1
ENST00000409764.1 ENST00000413447.1 |
CYCS
|
cytochrome c, somatic |
| chr11_+_18343800 | 0.09 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr2_-_202645835 | 0.09 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr1_-_151254362 | 0.09 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr22_-_36357671 | 0.09 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr6_+_126661253 | 0.08 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr1_-_154164534 | 0.08 |
ENST00000271850.7
ENST00000368530.2 |
TPM3
|
tropomyosin 3 |
| chrX_+_152240819 | 0.08 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr22_-_39096981 | 0.08 |
ENST00000427389.1
|
JOSD1
|
Josephin domain containing 1 |
| chr3_-_130745571 | 0.08 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr1_+_168148169 | 0.08 |
ENST00000367833.2
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr17_-_7155274 | 0.08 |
ENST00000318988.6
ENST00000575783.1 ENST00000573600.1 |
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr14_-_23446900 | 0.08 |
ENST00000556731.1
|
AJUBA
|
ajuba LIM protein |
| chr11_-_18343725 | 0.08 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr14_+_104095514 | 0.08 |
ENST00000348520.6
ENST00000380038.3 ENST00000389744.4 ENST00000557575.1 ENST00000553286.1 ENST00000347839.6 ENST00000555836.1 ENST00000334553.6 ENST00000246489.7 ENST00000557450.1 ENST00000452929.2 ENST00000554280.1 ENST00000445352.4 |
KLC1
|
kinesin light chain 1 |
| chr11_-_8680383 | 0.08 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr1_-_204165610 | 0.08 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr6_-_53213780 | 0.08 |
ENST00000304434.6
ENST00000370918.4 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr5_+_139055021 | 0.08 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr4_-_113558014 | 0.08 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chrX_-_152343394 | 0.08 |
ENST00000370261.1
|
PNMA6B
|
paraneoplastic Ma antigen family member 6B (pseudogene) |
| chr12_+_112856690 | 0.08 |
ENST00000392597.1
ENST00000351677.2 |
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr17_-_40540484 | 0.08 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr18_-_5540471 | 0.08 |
ENST00000581833.1
ENST00000544123.1 ENST00000342933.3 ENST00000400111.3 ENST00000585142.1 |
EPB41L3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr6_-_53213587 | 0.08 |
ENST00000542638.1
ENST00000370913.5 ENST00000541407.1 |
ELOVL5
|
ELOVL fatty acid elongase 5 |
| chr15_+_89905705 | 0.08 |
ENST00000560008.1
ENST00000561327.1 |
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr2_+_70142232 | 0.08 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr9_-_127533582 | 0.08 |
ENST00000416460.2
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr9_-_111696340 | 0.08 |
ENST00000374647.5
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr11_-_3818688 | 0.08 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr1_+_22379120 | 0.08 |
ENST00000400259.1
ENST00000344548.3 |
CDC42
|
cell division cycle 42 |
| chr1_+_28099700 | 0.08 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr6_-_10694766 | 0.08 |
ENST00000460742.2
ENST00000259983.3 ENST00000379586.1 |
C6orf52
|
chromosome 6 open reading frame 52 |
| chr7_-_124569864 | 0.08 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr1_+_25071848 | 0.08 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr5_+_138210919 | 0.08 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr17_+_74536164 | 0.08 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr8_+_38243951 | 0.08 |
ENST00000297720.5
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr5_+_172332220 | 0.08 |
ENST00000518247.1
ENST00000326654.2 |
ERGIC1
|
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
| chr2_-_151395525 | 0.08 |
ENST00000439275.1
|
RND3
|
Rho family GTPase 3 |
| chr5_+_159343688 | 0.08 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr19_+_2841433 | 0.07 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chrX_+_12993202 | 0.07 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr8_-_133772870 | 0.07 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr17_+_4643300 | 0.07 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr14_+_32030582 | 0.07 |
ENST00000550649.1
ENST00000281081.7 |
NUBPL
|
nucleotide binding protein-like |
| chrX_+_12993336 | 0.07 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr17_-_58603482 | 0.07 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr15_+_42841008 | 0.07 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr2_-_198175495 | 0.07 |
ENST00000409153.1
ENST00000409919.1 ENST00000539527.1 |
ANKRD44
|
ankyrin repeat domain 44 |
| chr3_+_185080908 | 0.07 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr2_+_152214098 | 0.07 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr5_-_172198190 | 0.07 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr14_+_35515582 | 0.07 |
ENST00000382406.3
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr13_+_111767650 | 0.07 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr3_-_21792838 | 0.07 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr1_+_110527308 | 0.07 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr17_-_7155802 | 0.07 |
ENST00000572043.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr12_+_56511943 | 0.07 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr11_+_18416133 | 0.07 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr4_-_76598544 | 0.07 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr17_+_4643337 | 0.07 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr17_+_4802713 | 0.07 |
ENST00000521575.1
ENST00000381365.3 |
C17orf107
|
chromosome 17 open reading frame 107 |
| chr11_+_18416103 | 0.07 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr11_+_18344106 | 0.07 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr17_+_41177220 | 0.07 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr21_-_40720995 | 0.07 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr19_-_9649253 | 0.07 |
ENST00000593003.1
|
ZNF426
|
zinc finger protein 426 |
| chr9_-_70490107 | 0.07 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr6_-_121655552 | 0.07 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr8_+_98788003 | 0.07 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr2_+_114195268 | 0.07 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr12_-_31881944 | 0.07 |
ENST00000537562.1
ENST00000537960.1 ENST00000536761.1 ENST00000542781.1 ENST00000457428.2 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr12_-_31882027 | 0.07 |
ENST00000541931.1
ENST00000535408.1 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr1_-_22109682 | 0.07 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr12_-_5352315 | 0.07 |
ENST00000536518.1
|
RP11-319E16.1
|
RP11-319E16.1 |
| chr19_-_44100275 | 0.06 |
ENST00000422989.1
ENST00000598324.1 |
IRGQ
|
immunity-related GTPase family, Q |
| chr2_-_61389168 | 0.06 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB5_CREM_JUNB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.2 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.8 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.3 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0018963 | phthalate metabolic process(GO:0018963) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:1990180 | mitochondrial tRNA 3'-end processing(GO:1990180) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.1 | GO:0021896 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.3 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.0 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.4 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0039007 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.0 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.0 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |