|
chr19_+_50031547
Show fit
|
0.86 |
ENST00000597801.1
|
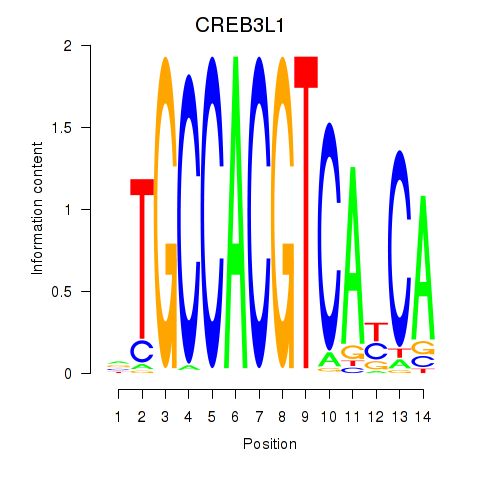
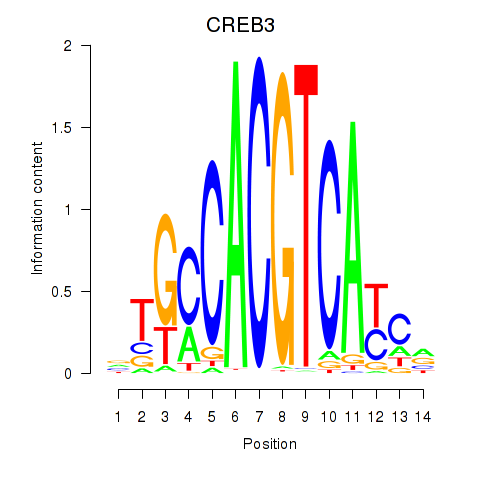
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr6_+_31916733
Show fit
|
0.47 |
ENST00000483004.1
|
CFB
|
complement factor B
|
|
chr6_+_32936942
Show fit
|
0.42 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2
|
|
chr22_+_38864041
Show fit
|
0.40 |
ENST00000216014.4
ENST00000409006.3
|
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3
|
|
chr5_+_68389807
Show fit
|
0.36 |
ENST00000380860.4
ENST00000504103.1
ENST00000502979.1
|
SLC30A5
|
solute carrier family 30 (zinc transporter), member 5
|
|
chr4_+_165675197
Show fit
|
0.31 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2
|
|
chr14_+_24600484
Show fit
|
0.29 |
ENST00000267426.5
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr3_-_57583185
Show fit
|
0.27 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr12_-_105352047
Show fit
|
0.27 |
ENST00000432951.1
ENST00000415674.1
ENST00000424946.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2
|
|
chr19_+_10527449
Show fit
|
0.26 |
ENST00000592685.1
ENST00000380702.2
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr11_+_125495862
Show fit
|
0.26 |
ENST00000428830.2
ENST00000544373.1
ENST00000527013.1
ENST00000526937.1
ENST00000534685.1
|
CHEK1
|
checkpoint kinase 1
|
|
chr12_-_122750957
Show fit
|
0.24 |
ENST00000451053.2
|
VPS33A
|
vacuolar protein sorting 33 homolog A (S. cerevisiae)
|
|
chr22_-_28392227
Show fit
|
0.24 |
ENST00000431039.1
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr8_-_95220775
Show fit
|
0.21 |
ENST00000441892.2
ENST00000521491.1
ENST00000027335.3
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine)
|
|
chr1_-_119682812
Show fit
|
0.19 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial
|
|
chr15_+_28623784
Show fit
|
0.19 |
ENST00000526619.2
ENST00000337838.7
ENST00000532622.2
|
GOLGA8F
|
golgin A8 family, member F
|
|
chr6_+_26204825
Show fit
|
0.17 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e
|
|
chr8_+_22423479
Show fit
|
0.17 |
ENST00000522721.1
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr15_-_28778117
Show fit
|
0.16 |
ENST00000525590.2
ENST00000329523.6
|
GOLGA8G
|
golgin A8 family, member G
|
|
chr6_-_90062543
Show fit
|
0.16 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1
|
|
chr3_-_57583130
Show fit
|
0.16 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr15_+_23599993
Show fit
|
0.16 |
ENST00000562295.1
|
GOLGA8S
|
golgin A8 family, member S
|
|
chr8_-_29208183
Show fit
|
0.16 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr12_+_113376249
Show fit
|
0.15 |
ENST00000551007.1
ENST00000548514.1
|
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa
|
|
chr3_+_121554046
Show fit
|
0.15 |
ENST00000273668.2
ENST00000451944.2
|
EAF2
|
ELL associated factor 2
|
|
chrX_+_101854426
Show fit
|
0.15 |
ENST00000536530.1
ENST00000473968.1
ENST00000604957.1
ENST00000477663.1
ENST00000479502.1
|
ARMCX5
|
armadillo repeat containing, X-linked 5
|
|
chr1_-_202311088
Show fit
|
0.15 |
ENST00000367274.4
|
UBE2T
|
ubiquitin-conjugating enzyme E2T (putative)
|
|
chr2_-_242211359
Show fit
|
0.14 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein
|
|
chrX_+_134124968
Show fit
|
0.14 |
ENST00000330288.4
|
SMIM10
|
small integral membrane protein 10
|
|
chr4_-_83351005
Show fit
|
0.14 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr6_+_32937083
Show fit
|
0.14 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2
|
|
chr7_+_128379449
Show fit
|
0.13 |
ENST00000479257.1
|
CALU
|
calumenin
|
|
chr17_+_57642886
Show fit
|
0.13 |
ENST00000251241.4
ENST00000451169.2
ENST00000425628.3
ENST00000584385.1
ENST00000580030.1
|
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr1_+_162531294
Show fit
|
0.13 |
ENST00000367926.4
ENST00000271469.3
|
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1
|
|
chr16_-_11692320
Show fit
|
0.13 |
ENST00000571627.1
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr2_-_99917639
Show fit
|
0.13 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1
|
|
chr2_-_202645612
Show fit
|
0.13 |
ENST00000409632.2
ENST00000410052.1
ENST00000467448.1
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr11_+_125496124
Show fit
|
0.13 |
ENST00000533778.2
ENST00000534070.1
|
CHEK1
|
checkpoint kinase 1
|
|
chr11_-_36310958
Show fit
|
0.12 |
ENST00000532705.1
ENST00000263401.5
ENST00000452374.2
|
COMMD9
|
COMM domain containing 9
|
|
chr10_-_22292675
Show fit
|
0.12 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1
|
|
chr12_-_56321397
Show fit
|
0.12 |
ENST00000557259.1
ENST00000549939.1
|
WIBG
|
within bgcn homolog (Drosophila)
|
|
chr2_-_172967621
Show fit
|
0.12 |
ENST00000234198.4
ENST00000466293.2
|
DLX2
|
distal-less homeobox 2
|
|
chr5_+_176730769
Show fit
|
0.12 |
ENST00000303204.4
ENST00000503216.1
|
PRELID1
|
PRELI domain containing 1
|
|
chr12_-_56320887
Show fit
|
0.11 |
ENST00000398213.4
|
WIBG
|
within bgcn homolog (Drosophila)
|
|
chr10_-_46089939
Show fit
|
0.11 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase
|
|
chr17_-_6554747
Show fit
|
0.11 |
ENST00000574128.1
|
MED31
|
mediator complex subunit 31
|
|
chr15_-_74988281
Show fit
|
0.11 |
ENST00000566828.1
ENST00000563009.1
ENST00000568176.1
ENST00000566243.1
ENST00000566219.1
ENST00000426797.3
ENST00000566119.1
ENST00000315127.4
|
EDC3
|
enhancer of mRNA decapping 3
|
|
chr6_+_151773583
Show fit
|
0.11 |
ENST00000545879.1
|
C6orf211
|
chromosome 6 open reading frame 211
|
|
chr15_-_23692381
Show fit
|
0.11 |
ENST00000567107.1
ENST00000345070.5
ENST00000312015.5
|
GOLGA6L2
|
golgin A6 family-like 2
|
|
chr6_+_24775641
Show fit
|
0.11 |
ENST00000378054.2
ENST00000476555.1
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr17_+_33307503
Show fit
|
0.11 |
ENST00000378526.4
ENST00000585941.1
ENST00000262327.5
ENST00000592690.1
ENST00000585740.1
|
LIG3
|
ligase III, DNA, ATP-dependent
|
|
chr20_+_18118486
Show fit
|
0.11 |
ENST00000432901.3
|
PET117
|
PET117 homolog (S. cerevisiae)
|
|
chr12_+_1100423
Show fit
|
0.11 |
ENST00000592048.1
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr11_-_62341445
Show fit
|
0.10 |
ENST00000329251.4
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma
|
|
chr15_+_40987327
Show fit
|
0.10 |
ENST00000423169.2
ENST00000267868.3
ENST00000557850.1
ENST00000532743.1
ENST00000382643.3
|
RAD51
|
RAD51 recombinase
|
|
chr8_+_109455830
Show fit
|
0.10 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2
|
|
chrX_-_19689106
Show fit
|
0.10 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr12_-_56321649
Show fit
|
0.10 |
ENST00000454792.2
ENST00000408946.2
|
WIBG
|
within bgcn homolog (Drosophila)
|
|
chr22_+_50981079
Show fit
|
0.10 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34
|
|
chr3_-_118959733
Show fit
|
0.10 |
ENST00000459778.1
ENST00000359213.3
|
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4
|
|
chr9_+_100818976
Show fit
|
0.10 |
ENST00000210444.5
|
NANS
|
N-acetylneuraminic acid synthase
|
|
chr1_+_249200395
Show fit
|
0.10 |
ENST00000355360.4
ENST00000329291.5
ENST00000539153.1
|
PGBD2
|
piggyBac transposable element derived 2
|
|
chr4_+_6202448
Show fit
|
0.09 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1
|
|
chrX_-_102941596
Show fit
|
0.09 |
ENST00000441076.2
ENST00000422355.1
ENST00000442614.1
ENST00000422154.2
ENST00000451301.1
|
MORF4L2
|
mortality factor 4 like 2
|
|
chr3_-_25824353
Show fit
|
0.09 |
ENST00000427041.1
|
NGLY1
|
N-glycanase 1
|
|
chr2_-_120980939
Show fit
|
0.09 |
ENST00000426077.2
|
TMEM185B
|
transmembrane protein 185B
|
|
chr3_-_57583052
Show fit
|
0.09 |
ENST00000496292.1
ENST00000489843.1
|
ARF4
|
ADP-ribosylation factor 4
|
|
chr6_+_32936353
Show fit
|
0.09 |
ENST00000374825.4
|
BRD2
|
bromodomain containing 2
|
|
chr12_-_31881944
Show fit
|
0.09 |
ENST00000537562.1
ENST00000537960.1
ENST00000536761.1
ENST00000542781.1
ENST00000457428.2
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr11_-_64646086
Show fit
|
0.09 |
ENST00000320631.3
|
EHD1
|
EH-domain containing 1
|
|
chr2_+_162165038
Show fit
|
0.09 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14
|
|
chr2_-_69614373
Show fit
|
0.09 |
ENST00000361060.5
ENST00000357308.4
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr14_-_50087312
Show fit
|
0.09 |
ENST00000298289.6
|
RPL36AL
|
ribosomal protein L36a-like
|
|
chrX_-_153059811
Show fit
|
0.09 |
ENST00000427365.2
ENST00000444450.1
ENST00000370093.1
|
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma
|
|
chr2_-_27341966
Show fit
|
0.09 |
ENST00000402394.1
ENST00000402550.1
ENST00000260595.5
|
CGREF1
|
cell growth regulator with EF-hand domain 1
|
|
chr14_+_68086515
Show fit
|
0.09 |
ENST00000261783.3
|
ARG2
|
arginase 2
|
|
chr12_-_93835665
Show fit
|
0.09 |
ENST00000552442.1
ENST00000550657.1
|
UBE2N
|
ubiquitin-conjugating enzyme E2N
|
|
chr12_-_49318715
Show fit
|
0.09 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa
|
|
chr16_+_68057179
Show fit
|
0.09 |
ENST00000567100.1
ENST00000432752.1
ENST00000569289.1
ENST00000564781.1
|
DUS2
|
dihydrouridine synthase 2
|
|
chr8_-_38126675
Show fit
|
0.09 |
ENST00000531823.1
ENST00000534339.1
ENST00000524616.1
ENST00000422581.2
ENST00000424479.2
ENST00000419686.2
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B
|
|
chr3_-_25824872
Show fit
|
0.09 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1
|
|
chr12_+_56110247
Show fit
|
0.08 |
ENST00000551926.1
|
BLOC1S1
|
biogenesis of lysosomal organelles complex-1, subunit 1
|
|
chr3_-_33759699
Show fit
|
0.08 |
ENST00000399362.4
ENST00000359576.5
ENST00000307312.7
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr2_-_202645835
Show fit
|
0.08 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile)
|
|
chr3_-_139108475
Show fit
|
0.08 |
ENST00000515006.1
ENST00000513274.1
ENST00000514508.1
ENST00000507777.1
ENST00000512153.1
ENST00000333188.5
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime)
|
|
chr20_+_23331373
Show fit
|
0.08 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1
|
|
chr12_-_76953453
Show fit
|
0.08 |
ENST00000549570.1
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr12_+_1100370
Show fit
|
0.08 |
ENST00000543086.3
ENST00000546231.2
ENST00000397203.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr1_+_228645796
Show fit
|
0.08 |
ENST00000369160.2
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr1_+_222913009
Show fit
|
0.08 |
ENST00000456298.1
|
FAM177B
|
family with sequence similarity 177, member B
|
|
chr4_+_128982490
Show fit
|
0.08 |
ENST00000394288.3
ENST00000432347.2
ENST00000264584.5
ENST00000441387.1
ENST00000427266.1
ENST00000354456.3
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr7_+_140396946
Show fit
|
0.08 |
ENST00000476470.1
ENST00000471136.1
|
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa
|
|
chr4_+_128982430
Show fit
|
0.08 |
ENST00000512292.1
ENST00000508819.1
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr6_+_116692102
Show fit
|
0.08 |
ENST00000359564.2
|
DSE
|
dermatan sulfate epimerase
|
|
chr9_+_112542572
Show fit
|
0.08 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough
|
|
chr3_+_3168600
Show fit
|
0.08 |
ENST00000251607.6
ENST00000339437.6
ENST00000280591.6
ENST00000420393.1
|
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1
|
|
chr7_+_30174668
Show fit
|
0.08 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus)
|
|
chr17_-_6554877
Show fit
|
0.08 |
ENST00000225728.3
ENST00000575197.1
|
MED31
|
mediator complex subunit 31
|
|
chr12_+_1100449
Show fit
|
0.08 |
ENST00000360905.4
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr6_-_101329157
Show fit
|
0.08 |
ENST00000369143.2
|
ASCC3
|
activating signal cointegrator 1 complex subunit 3
|
|
chr17_-_61777090
Show fit
|
0.08 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2
|
|
chrX_-_118699325
Show fit
|
0.07 |
ENST00000320339.4
ENST00000371594.4
ENST00000536133.1
|
CXorf56
|
chromosome X open reading frame 56
|
|
chr1_+_11866270
Show fit
|
0.07 |
ENST00000376497.3
ENST00000376487.3
ENST00000376496.3
|
CLCN6
|
chloride channel, voltage-sensitive 6
|
|
chr16_-_57219721
Show fit
|
0.07 |
ENST00000562406.1
ENST00000568671.1
ENST00000567044.1
|
FAM192A
|
family with sequence similarity 192, member A
|
|
chr2_+_96068436
Show fit
|
0.07 |
ENST00000445649.1
ENST00000447036.1
ENST00000233379.4
ENST00000418606.1
|
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A
|
|
chr8_+_22422749
Show fit
|
0.07 |
ENST00000523900.1
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr10_-_90342947
Show fit
|
0.07 |
ENST00000437752.1
ENST00000331772.4
|
RNLS
|
renalase, FAD-dependent amine oxidase
|
|
chr19_-_41256207
Show fit
|
0.07 |
ENST00000598485.2
ENST00000470681.1
ENST00000339153.3
ENST00000598729.1
|
C19orf54
|
chromosome 19 open reading frame 54
|
|
chr3_+_100211412
Show fit
|
0.07 |
ENST00000323523.4
ENST00000403410.1
ENST00000449609.1
|
TMEM45A
|
transmembrane protein 45A
|
|
chr10_+_35484793
Show fit
|
0.07 |
ENST00000488741.1
ENST00000474931.1
ENST00000468236.1
ENST00000344351.5
ENST00000490511.1
|
CREM
|
cAMP responsive element modulator
|
|
chr5_-_1799932
Show fit
|
0.07 |
ENST00000382647.7
ENST00000505059.2
|
MRPL36
|
mitochondrial ribosomal protein L36
|
|
chr3_-_132378919
Show fit
|
0.07 |
ENST00000355458.3
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11
|
|
chr17_-_191188
Show fit
|
0.07 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr5_+_133984462
Show fit
|
0.07 |
ENST00000398844.2
ENST00000322887.4
|
SEC24A
|
SEC24 family member A
|
|
chr5_-_1799965
Show fit
|
0.07 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36
|
|
chr1_+_222791417
Show fit
|
0.07 |
ENST00000344922.5
ENST00000344441.6
ENST00000344507.1
|
MIA3
|
melanoma inhibitory activity family, member 3
|
|
chr1_+_45205478
Show fit
|
0.07 |
ENST00000452259.1
ENST00000372224.4
|
KIF2C
|
kinesin family member 2C
|
|
chr20_+_18118703
Show fit
|
0.07 |
ENST00000464792.1
|
CSRP2BP
|
CSRP2 binding protein
|
|
chr4_+_128982416
Show fit
|
0.07 |
ENST00000326639.6
|
LARP1B
|
La ribonucleoprotein domain family, member 1B
|
|
chr5_-_10761206
Show fit
|
0.07 |
ENST00000432074.2
ENST00000230895.6
|
DAP
|
death-associated protein
|
|
chr22_-_20850070
Show fit
|
0.07 |
ENST00000440659.2
ENST00000458248.1
ENST00000443285.1
ENST00000444967.1
ENST00000451553.1
ENST00000431430.1
|
KLHL22
|
kelch-like family member 22
|
|
chr6_-_74231303
Show fit
|
0.07 |
ENST00000309268.6
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr3_+_3168692
Show fit
|
0.07 |
ENST00000402675.1
ENST00000413000.1
|
TRNT1
|
tRNA nucleotidyl transferase, CCA-adding, 1
|
|
chr3_-_119182453
Show fit
|
0.07 |
ENST00000491685.1
ENST00000461654.1
|
TMEM39A
|
transmembrane protein 39A
|
|
chr22_-_24093267
Show fit
|
0.07 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70
|
|
chr16_+_31128978
Show fit
|
0.07 |
ENST00000448516.2
ENST00000219797.4
|
KAT8
|
K(lysine) acetyltransferase 8
|
|
chr3_+_28390637
Show fit
|
0.07 |
ENST00000420223.1
ENST00000383768.2
|
ZCWPW2
|
zinc finger, CW type with PWWP domain 2
|
|
chr4_-_83350580
Show fit
|
0.06 |
ENST00000349655.4
ENST00000602300.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr2_-_43823119
Show fit
|
0.06 |
ENST00000403856.1
ENST00000404790.1
ENST00000405975.2
ENST00000415080.2
|
THADA
|
thyroid adenoma associated
|
|
chr19_-_54619006
Show fit
|
0.06 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr5_-_39424961
Show fit
|
0.06 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila)
|
|
chr6_-_74231444
Show fit
|
0.06 |
ENST00000331523.2
ENST00000356303.2
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr1_-_38156153
Show fit
|
0.06 |
ENST00000464085.1
ENST00000486637.1
ENST00000358011.4
ENST00000461359.1
|
C1orf109
|
chromosome 1 open reading frame 109
|
|
chr7_+_23145366
Show fit
|
0.06 |
ENST00000339077.5
ENST00000322275.5
ENST00000539124.1
ENST00000542558.1
|
KLHL7
|
kelch-like family member 7
|
|
chr20_+_57467204
Show fit
|
0.06 |
ENST00000603546.1
|
GNAS
|
GNAS complex locus
|
|
chr7_-_73184588
Show fit
|
0.06 |
ENST00000395145.2
|
CLDN3
|
claudin 3
|
|
chr3_+_133292574
Show fit
|
0.06 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr1_-_43833628
Show fit
|
0.06 |
ENST00000413844.2
ENST00000372458.3
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr19_+_5681011
Show fit
|
0.06 |
ENST00000581893.1
ENST00000411793.2
ENST00000301382.4
ENST00000581773.1
ENST00000423665.2
ENST00000583928.1
ENST00000342970.2
ENST00000422535.2
ENST00000581521.1
ENST00000339423.2
|
HSD11B1L
|
hydroxysteroid (11-beta) dehydrogenase 1-like
|
|
chr16_+_57220049
Show fit
|
0.06 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1
|
|
chr3_+_134205000
Show fit
|
0.06 |
ENST00000512894.1
ENST00000513612.2
ENST00000606977.1
|
CEP63
|
centrosomal protein 63kDa
|
|
chr11_-_2906979
Show fit
|
0.06 |
ENST00000380725.1
ENST00000313407.6
ENST00000430149.2
ENST00000440480.2
ENST00000414822.3
|
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2)
|
|
chr1_+_41249539
Show fit
|
0.06 |
ENST00000347132.5
ENST00000509682.2
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr2_-_128643496
Show fit
|
0.06 |
ENST00000272647.5
|
AMMECR1L
|
AMMECR1-like
|
|
chrX_+_16141667
Show fit
|
0.06 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor
|
|
chr12_-_31882027
Show fit
|
0.06 |
ENST00000541931.1
ENST00000535408.1
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae)
|
|
chr6_+_44355257
Show fit
|
0.06 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like
|
|
chr22_+_39916558
Show fit
|
0.06 |
ENST00000337304.2
ENST00000396680.1
|
ATF4
|
activating transcription factor 4
|
|
chr16_+_68056844
Show fit
|
0.06 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2
|
|
chr1_+_1567474
Show fit
|
0.06 |
ENST00000356026.5
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr15_+_40987661
Show fit
|
0.06 |
ENST00000526763.1
|
RAD51
|
RAD51 recombinase
|
|
chr18_-_47018869
Show fit
|
0.05 |
ENST00000583036.1
ENST00000580261.1
|
RPL17
|
ribosomal protein L17
|
|
chr3_-_28390415
Show fit
|
0.05 |
ENST00000414162.1
ENST00000420543.2
|
AZI2
|
5-azacytidine induced 2
|
|
chr9_-_98079965
Show fit
|
0.05 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C
|
|
chr3_-_33759541
Show fit
|
0.05 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2
|
|
chr9_+_117350009
Show fit
|
0.05 |
ENST00000374050.3
|
ATP6V1G1
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1
|
|
chr5_-_150138246
Show fit
|
0.05 |
ENST00000518015.1
|
DCTN4
|
dynactin 4 (p62)
|
|
chr17_-_48450265
Show fit
|
0.05 |
ENST00000507088.1
|
MRPL27
|
mitochondrial ribosomal protein L27
|
|
chr18_-_47017956
Show fit
|
0.05 |
ENST00000584895.1
ENST00000332968.6
ENST00000580210.1
ENST00000579408.1
|
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough
ribosomal protein L17
|
|
chr6_+_24775153
Show fit
|
0.05 |
ENST00000356509.3
ENST00000230056.3
|
GMNN
|
geminin, DNA replication inhibitor
|
|
chr12_-_125398850
Show fit
|
0.05 |
ENST00000535859.1
ENST00000546271.1
ENST00000540700.1
ENST00000546120.1
|
UBC
|
ubiquitin C
|
|
chr20_-_1306351
Show fit
|
0.05 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chrX_+_8433376
Show fit
|
0.05 |
ENST00000440654.2
ENST00000381029.4
|
VCX3B
|
variable charge, X-linked 3B
|
|
chr16_-_11836595
Show fit
|
0.05 |
ENST00000356957.3
ENST00000283033.5
|
TXNDC11
|
thioredoxin domain containing 11
|
|
chr19_+_55476620
Show fit
|
0.05 |
ENST00000543010.1
ENST00000391721.4
ENST00000339757.7
|
NLRP2
|
NLR family, pyrin domain containing 2
|
|
chr13_-_52026730
Show fit
|
0.05 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6
|
|
chr6_+_151561085
Show fit
|
0.05 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr2_+_27805880
Show fit
|
0.05 |
ENST00000379717.1
ENST00000355467.4
ENST00000556601.1
ENST00000416005.2
|
ZNF512
|
zinc finger protein 512
|
|
chr17_-_48450534
Show fit
|
0.05 |
ENST00000503633.1
ENST00000442592.3
ENST00000225969.4
|
MRPL27
|
mitochondrial ribosomal protein L27
|
|
chr19_-_54618650
Show fit
|
0.05 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia)
|
|
chr2_-_33824336
Show fit
|
0.05 |
ENST00000431950.1
ENST00000403368.1
ENST00000441530.2
|
FAM98A
|
family with sequence similarity 98, member A
|
|
chr1_-_11866034
Show fit
|
0.05 |
ENST00000376590.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H)
|
|
chr13_-_31040060
Show fit
|
0.05 |
ENST00000326004.4
ENST00000341423.5
|
HMGB1
|
high mobility group box 1
|
|
chr16_+_68057153
Show fit
|
0.05 |
ENST00000358896.6
ENST00000568099.2
|
DUS2
|
dihydrouridine synthase 2
|
|
chr3_-_118959716
Show fit
|
0.05 |
ENST00000467604.1
ENST00000491906.1
ENST00000475803.1
ENST00000479150.1
ENST00000470111.1
ENST00000459820.1
|
B4GALT4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4
|
|
chr2_-_43823093
Show fit
|
0.05 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated
|
|
chr11_-_118927561
Show fit
|
0.05 |
ENST00000530473.1
|
HYOU1
|
hypoxia up-regulated 1
|
|
chr12_+_10365404
Show fit
|
0.05 |
ENST00000266458.5
ENST00000421801.2
ENST00000544284.1
ENST00000545047.1
ENST00000543602.1
ENST00000545887.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr3_+_50284321
Show fit
|
0.05 |
ENST00000451956.1
|
GNAI2
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2
|
|
chr1_+_45205498
Show fit
|
0.05 |
ENST00000372218.4
|
KIF2C
|
kinesin family member 2C
|
|
chr2_+_27255806
Show fit
|
0.05 |
ENST00000238788.9
ENST00000404032.3
|
TMEM214
|
transmembrane protein 214
|
|
chr2_+_233562015
Show fit
|
0.05 |
ENST00000427233.1
ENST00000373566.3
ENST00000373563.4
ENST00000428883.1
ENST00000456491.1
ENST00000409480.1
ENST00000421433.1
ENST00000425040.1
ENST00000430720.1
ENST00000409547.1
ENST00000423659.1
ENST00000409196.3
ENST00000409451.3
ENST00000429187.1
ENST00000440945.1
|
GIGYF2
|
GRB10 interacting GYF protein 2
|
|
chr2_-_98280383
Show fit
|
0.05 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast)
|
|
chr12_-_51566849
Show fit
|
0.05 |
ENST00000549867.1
ENST00000307660.4
|
TFCP2
|
transcription factor CP2
|
|
chr2_+_192542879
Show fit
|
0.05 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1
|
|
chr3_+_128598433
Show fit
|
0.04 |
ENST00000308982.7
ENST00000514336.1
|
ACAD9
|
acyl-CoA dehydrogenase family, member 9
|
|
chr4_+_75023816
Show fit
|
0.04 |
ENST00000395759.2
ENST00000331145.6
ENST00000359107.5
ENST00000325278.6
|
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like
|
|
chr1_-_32110467
Show fit
|
0.04 |
ENST00000440872.2
ENST00000373703.4
|
PEF1
|
penta-EF-hand domain containing 1
|
|
chr20_-_1306391
Show fit
|
0.04 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2
|
|
chr19_+_41256764
Show fit
|
0.04 |
ENST00000243563.3
ENST00000601253.1
ENST00000597353.1
ENST00000599362.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A
|
|
chr14_+_103995503
Show fit
|
0.04 |
ENST00000389749.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae)
|
|
chr11_-_3818688
Show fit
|
0.04 |
ENST00000355260.3
ENST00000397004.4
ENST00000397007.4
ENST00000532475.1
|
NUP98
|
nucleoporin 98kDa
|
|
chr3_-_28389922
Show fit
|
0.04 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2
|
|
chrX_-_6453159
Show fit
|
0.04 |
ENST00000381089.3
ENST00000398729.1
|
VCX3A
|
variable charge, X-linked 3A
|
|
chr6_+_127588020
Show fit
|
0.04 |
ENST00000309649.3
ENST00000610162.1
ENST00000610153.1
ENST00000608991.1
ENST00000480444.1
|
RNF146
|
ring finger protein 146
|
|
chr15_+_44084503
Show fit
|
0.04 |
ENST00000409960.2
ENST00000409646.1
ENST00000594896.1
ENST00000339624.5
ENST00000409291.1
ENST00000402131.1
ENST00000403425.1
ENST00000430901.1
|
SERF2
|
small EDRK-rich factor 2
|
|
chr11_-_118927816
Show fit
|
0.04 |
ENST00000534233.1
ENST00000532752.1
ENST00000525859.1
ENST00000404233.3
ENST00000532421.1
ENST00000543287.1
ENST00000527310.2
ENST00000529972.1
|
HYOU1
|
hypoxia up-regulated 1
|
|
chrX_-_7895479
Show fit
|
0.04 |
ENST00000381042.4
|
PNPLA4
|
patatin-like phospholipase domain containing 4
|
|
chr12_-_125399573
Show fit
|
0.04 |
ENST00000339647.5
|
UBC
|
ubiquitin C
|
|
chr12_+_56552128
Show fit
|
0.04 |
ENST00000548580.1
ENST00000293422.5
ENST00000348108.4
ENST00000549017.1
ENST00000549566.1
ENST00000536128.1
ENST00000547649.1
ENST00000547408.1
ENST00000551589.1
ENST00000549392.1
ENST00000548400.1
ENST00000548293.1
|
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle
|