Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
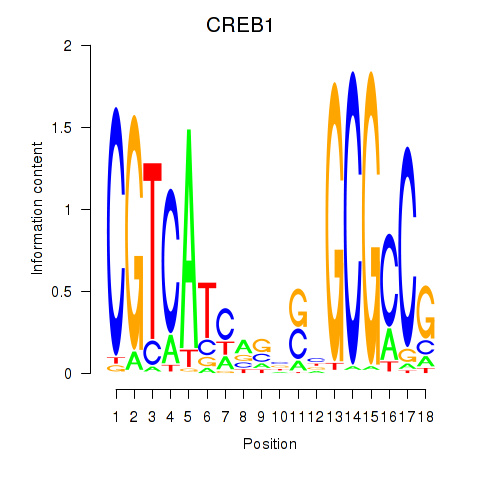
Results for CREB1
Z-value: 1.10
Transcription factors associated with CREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB1
|
ENSG00000118260.10 | cAMP responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB1 | hg19_v2_chr2_+_208414985_208415079 | -0.69 | 1.3e-01 | Click! |
Activity profile of CREB1 motif
Sorted Z-values of CREB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_17103951 | 1.59 |
ENST00000520178.1
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr1_-_1590418 | 0.69 |
ENST00000341028.7
|
CDK11B
|
cyclin-dependent kinase 11B |
| chr5_-_33297946 | 0.53 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr13_+_36920569 | 0.51 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr8_+_17104401 | 0.50 |
ENST00000324815.3
ENST00000518038.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr16_-_25122785 | 0.48 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr16_+_19078911 | 0.43 |
ENST00000321998.5
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr16_-_25122735 | 0.42 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr9_-_135754164 | 0.37 |
ENST00000298545.3
|
AK8
|
adenylate kinase 8 |
| chr4_+_76439649 | 0.36 |
ENST00000507557.1
|
THAP6
|
THAP domain containing 6 |
| chr5_-_72861175 | 0.33 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr3_-_196295437 | 0.33 |
ENST00000429115.1
|
WDR53
|
WD repeat domain 53 |
| chr21_-_44299626 | 0.33 |
ENST00000330317.2
ENST00000398208.2 |
WDR4
|
WD repeat domain 4 |
| chr3_-_196295468 | 0.32 |
ENST00000332629.5
ENST00000433160.1 |
WDR53
|
WD repeat domain 53 |
| chr6_-_170151603 | 0.31 |
ENST00000366774.3
|
TCTE3
|
t-complex-associated-testis-expressed 3 |
| chr2_-_27579623 | 0.31 |
ENST00000457748.1
|
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr4_-_146019693 | 0.30 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr4_-_146019335 | 0.29 |
ENST00000451299.2
ENST00000507656.1 ENST00000309439.5 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr16_+_19079311 | 0.28 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr4_-_8442438 | 0.28 |
ENST00000356406.5
ENST00000413009.2 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr3_-_119182506 | 0.27 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr17_-_58469591 | 0.27 |
ENST00000589335.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr16_+_68057179 | 0.26 |
ENST00000567100.1
ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2
|
dihydrouridine synthase 2 |
| chr8_-_145742862 | 0.25 |
ENST00000524998.1
|
RECQL4
|
RecQ protein-like 4 |
| chr3_+_45017722 | 0.25 |
ENST00000265564.7
|
EXOSC7
|
exosome component 7 |
| chr15_+_41186637 | 0.25 |
ENST00000558474.1
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr1_-_186344802 | 0.25 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr11_-_64889649 | 0.25 |
ENST00000434372.2
|
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr22_+_26879817 | 0.24 |
ENST00000215917.7
|
SRRD
|
SRR1 domain containing |
| chr14_+_105219437 | 0.24 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr3_+_11313995 | 0.23 |
ENST00000451513.1
ENST00000435760.1 ENST00000451830.1 |
ATG7
|
autophagy related 7 |
| chr15_-_90198659 | 0.23 |
ENST00000394412.3
|
KIF7
|
kinesin family member 7 |
| chr19_-_54618650 | 0.23 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr8_+_24772455 | 0.21 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr9_-_123555655 | 0.21 |
ENST00000340778.5
ENST00000453291.1 ENST00000608872.1 |
FBXW2
|
F-box and WD repeat domain containing 2 |
| chr1_-_245134273 | 0.21 |
ENST00000607453.1
|
RP11-156E8.1
|
Uncharacterized protein |
| chr9_+_140083099 | 0.21 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr17_-_47022140 | 0.21 |
ENST00000290330.3
|
SNF8
|
SNF8, ESCRT-II complex subunit |
| chr16_+_68057153 | 0.19 |
ENST00000358896.6
ENST00000568099.2 |
DUS2
|
dihydrouridine synthase 2 |
| chr12_+_54393880 | 0.19 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr2_-_21022818 | 0.19 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr2_-_220110111 | 0.19 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr12_-_58165870 | 0.19 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr3_+_141457105 | 0.19 |
ENST00000480908.1
ENST00000393000.3 |
RNF7
|
ring finger protein 7 |
| chr1_-_53704157 | 0.19 |
ENST00000371466.4
ENST00000371470.3 |
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr16_-_3767551 | 0.18 |
ENST00000246957.5
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr8_-_10697370 | 0.18 |
ENST00000314787.3
ENST00000426190.2 ENST00000519088.1 |
PINX1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr12_+_108079664 | 0.18 |
ENST00000541166.1
|
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr3_-_101405539 | 0.18 |
ENST00000469605.1
ENST00000495401.1 ENST00000394077.3 |
RPL24
|
ribosomal protein L24 |
| chr16_+_88636875 | 0.17 |
ENST00000569435.1
|
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr8_+_17104539 | 0.17 |
ENST00000521829.1
ENST00000521005.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr16_-_3767506 | 0.17 |
ENST00000538171.1
|
TRAP1
|
TNF receptor-associated protein 1 |
| chr16_+_25123041 | 0.17 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr8_-_10697281 | 0.17 |
ENST00000524114.1
ENST00000553390.1 ENST00000554914.1 |
PINX1
SOX7
SOX7
|
PIN2/TERF1 interacting, telomerase inhibitor 1 SRY (sex determining region Y)-box 7 Transcription factor SOX-7; Uncharacterized protein; cDNA FLJ58508, highly similar to Transcription factor SOX-7 |
| chr7_-_93633658 | 0.17 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr6_+_31939608 | 0.17 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr19_-_12807422 | 0.17 |
ENST00000380339.3
ENST00000544494.1 ENST00000393261.3 |
FBXW9
|
F-box and WD repeat domain containing 9 |
| chr12_+_56862301 | 0.17 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr2_+_234160340 | 0.16 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr4_+_146019421 | 0.16 |
ENST00000502586.1
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr16_+_2022036 | 0.16 |
ENST00000568546.1
|
TBL3
|
transducin (beta)-like 3 |
| chr6_-_32122106 | 0.16 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr16_-_790982 | 0.16 |
ENST00000301694.5
ENST00000251588.2 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr3_+_113775576 | 0.16 |
ENST00000485050.1
ENST00000281273.4 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr7_+_23145366 | 0.16 |
ENST00000339077.5
ENST00000322275.5 ENST00000539124.1 ENST00000542558.1 |
KLHL7
|
kelch-like family member 7 |
| chr5_+_72861560 | 0.15 |
ENST00000296792.4
ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr17_-_4607335 | 0.15 |
ENST00000570571.1
ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1
|
proline, glutamate and leucine rich protein 1 |
| chr8_-_126103948 | 0.15 |
ENST00000523297.1
|
KIAA0196
|
KIAA0196 |
| chr16_+_71879861 | 0.15 |
ENST00000427980.2
ENST00000568581.1 |
ATXN1L
IST1
|
ataxin 1-like increased sodium tolerance 1 homolog (yeast) |
| chr3_+_48488114 | 0.15 |
ENST00000421175.1
ENST00000320211.3 ENST00000346691.4 ENST00000357105.6 |
ATRIP
|
ATR interacting protein |
| chr20_+_3801162 | 0.15 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr16_+_68056844 | 0.15 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr10_-_119805558 | 0.15 |
ENST00000369199.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr1_+_12040238 | 0.15 |
ENST00000444836.1
ENST00000235329.5 |
MFN2
|
mitofusin 2 |
| chr22_+_20104947 | 0.14 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr2_+_101618965 | 0.14 |
ENST00000456292.1
ENST00000409711.1 ENST00000441435.1 |
RPL31
|
ribosomal protein L31 |
| chr7_+_44646177 | 0.14 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr3_-_52443799 | 0.14 |
ENST00000470173.1
ENST00000296288.5 |
BAP1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chrX_+_48660287 | 0.14 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr4_-_146019287 | 0.14 |
ENST00000502847.1
ENST00000513054.1 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr3_-_196669298 | 0.14 |
ENST00000411704.1
ENST00000452404.2 |
NCBP2
|
nuclear cap binding protein subunit 2, 20kDa |
| chr9_-_179018 | 0.13 |
ENST00000431099.2
ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1
|
COBW domain containing 1 |
| chr17_-_58469474 | 0.13 |
ENST00000300896.4
|
USP32
|
ubiquitin specific peptidase 32 |
| chr11_-_126138808 | 0.13 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr4_+_153457404 | 0.13 |
ENST00000604157.1
ENST00000594836.1 |
MIR4453
|
microRNA 4453 |
| chr6_-_31926208 | 0.13 |
ENST00000454913.1
ENST00000436289.2 |
NELFE
|
negative elongation factor complex member E |
| chr3_+_11314099 | 0.13 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr20_+_18488137 | 0.13 |
ENST00000450074.1
ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr6_-_8064567 | 0.13 |
ENST00000543936.1
ENST00000397457.2 |
BLOC1S5
|
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr1_+_202976493 | 0.13 |
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A |
| chr9_-_33001520 | 0.13 |
ENST00000463596.1
ENST00000379819.1 ENST00000397172.3 ENST00000379812.5 ENST00000477119.1 ENST00000379813.3 ENST00000379825.2 ENST00000309615.3 ENST00000476858.1 ENST00000473221.1 |
APTX
|
aprataxin |
| chr12_+_132628963 | 0.12 |
ENST00000330579.1
|
NOC4L
|
nucleolar complex associated 4 homolog (S. cerevisiae) |
| chr17_-_685559 | 0.12 |
ENST00000301329.6
|
GLOD4
|
glyoxalase domain containing 4 |
| chr7_-_150924121 | 0.12 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr5_-_133304473 | 0.12 |
ENST00000231512.3
|
C5orf15
|
chromosome 5 open reading frame 15 |
| chr17_+_29718642 | 0.12 |
ENST00000325874.8
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II) |
| chr22_+_41865109 | 0.12 |
ENST00000216254.4
ENST00000396512.3 |
ACO2
|
aconitase 2, mitochondrial |
| chr7_+_44646162 | 0.12 |
ENST00000439616.2
|
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr9_-_32573130 | 0.12 |
ENST00000350021.2
ENST00000379847.3 |
NDUFB6
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
| chr8_+_145743360 | 0.12 |
ENST00000527730.1
ENST00000529022.1 ENST00000292524.1 |
LRRC14
|
leucine rich repeat containing 14 |
| chr2_-_202316169 | 0.11 |
ENST00000430254.1
|
TRAK2
|
trafficking protein, kinesin binding 2 |
| chr20_+_3026591 | 0.11 |
ENST00000380325.3
|
MRPS26
|
mitochondrial ribosomal protein S26 |
| chr8_+_37594185 | 0.11 |
ENST00000518586.1
ENST00000335171.6 ENST00000521644.1 |
ERLIN2
|
ER lipid raft associated 2 |
| chrX_-_47518498 | 0.11 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr1_-_36107445 | 0.11 |
ENST00000373237.3
|
PSMB2
|
proteasome (prosome, macropain) subunit, beta type, 2 |
| chr22_+_20105012 | 0.11 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr15_-_73076030 | 0.11 |
ENST00000311669.8
|
ADPGK
|
ADP-dependent glucokinase |
| chr17_+_73663470 | 0.11 |
ENST00000583536.1
|
SAP30BP
|
SAP30 binding protein |
| chr19_+_10362577 | 0.11 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr1_-_92952433 | 0.11 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr8_-_29940569 | 0.11 |
ENST00000523761.1
|
TMEM66
|
transmembrane protein 66 |
| chr9_-_69262509 | 0.11 |
ENST00000377449.1
ENST00000382399.4 ENST00000377439.1 ENST00000377441.1 ENST00000377457.5 |
CBWD6
|
COBW domain containing 6 |
| chr21_-_46961716 | 0.11 |
ENST00000427839.1
|
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr17_-_685493 | 0.11 |
ENST00000536578.1
ENST00000301328.5 ENST00000576419.1 |
GLOD4
|
glyoxalase domain containing 4 |
| chr11_+_34127142 | 0.11 |
ENST00000257829.3
ENST00000531159.2 |
NAT10
|
N-acetyltransferase 10 (GCN5-related) |
| chr20_+_18488528 | 0.11 |
ENST00000377465.1
|
SEC23B
|
Sec23 homolog B (S. cerevisiae) |
| chr19_-_7694417 | 0.11 |
ENST00000358368.4
ENST00000534844.1 |
XAB2
|
XPA binding protein 2 |
| chr11_-_10562710 | 0.11 |
ENST00000528665.1
ENST00000265981.2 |
RNF141
|
ring finger protein 141 |
| chr2_+_96068436 | 0.10 |
ENST00000445649.1
ENST00000447036.1 ENST00000233379.4 ENST00000418606.1 |
FAHD2A
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr19_+_5720666 | 0.10 |
ENST00000381624.3
ENST00000381614.2 |
CATSPERD
|
catsper channel auxiliary subunit delta |
| chr17_-_8151353 | 0.10 |
ENST00000315684.8
|
CTC1
|
CTS telomere maintenance complex component 1 |
| chr12_+_108079509 | 0.10 |
ENST00000412830.3
ENST00000547995.1 |
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr2_+_114195268 | 0.10 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr3_+_196295482 | 0.10 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr1_-_32110467 | 0.10 |
ENST00000440872.2
ENST00000373703.4 |
PEF1
|
penta-EF-hand domain containing 1 |
| chr19_-_47987419 | 0.10 |
ENST00000536339.1
ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN
|
kaptin (actin binding protein) |
| chr16_+_29827832 | 0.10 |
ENST00000609618.1
|
PAGR1
|
PAXIP1-associated glutamate-rich protein 1 |
| chr19_-_16682987 | 0.10 |
ENST00000431408.1
ENST00000436553.2 ENST00000595753.1 |
SLC35E1
|
solute carrier family 35, member E1 |
| chr6_+_37400974 | 0.10 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr15_+_41186609 | 0.10 |
ENST00000220509.5
|
VPS18
|
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr16_+_29827285 | 0.10 |
ENST00000320330.6
|
PAGR1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr11_-_64570706 | 0.10 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr6_+_126661253 | 0.10 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr7_+_44646218 | 0.10 |
ENST00000444676.1
ENST00000222673.5 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr16_+_70332956 | 0.10 |
ENST00000288071.6
ENST00000393657.2 ENST00000355992.3 ENST00000567706.1 |
DDX19B
RP11-529K1.3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19B Uncharacterized protein |
| chr6_-_31940065 | 0.09 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr1_-_229644034 | 0.09 |
ENST00000366678.3
ENST00000261396.3 ENST00000537506.1 |
NUP133
|
nucleoporin 133kDa |
| chr10_+_103912137 | 0.09 |
ENST00000603742.1
ENST00000488254.2 ENST00000461421.1 ENST00000476468.1 ENST00000370007.5 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr3_+_128598433 | 0.09 |
ENST00000308982.7
ENST00000514336.1 |
ACAD9
|
acyl-CoA dehydrogenase family, member 9 |
| chr4_-_8430152 | 0.09 |
ENST00000514423.1
ENST00000503233.1 |
ACOX3
|
acyl-CoA oxidase 3, pristanoyl |
| chr3_+_141457030 | 0.09 |
ENST00000273480.3
|
RNF7
|
ring finger protein 7 |
| chrX_-_153285395 | 0.09 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr1_-_213031418 | 0.09 |
ENST00000356684.3
ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1
|
FLVCR1 antisense RNA 1 (head to head) |
| chr6_+_170151887 | 0.09 |
ENST00000588451.1
|
ERMARD
|
ER membrane-associated RNA degradation |
| chr7_-_1177874 | 0.09 |
ENST00000397098.3
ENST00000357429.6 ENST00000397100.2 ENST00000491163.1 |
C7orf50
|
chromosome 7 open reading frame 50 |
| chr12_+_69633317 | 0.09 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr22_-_41864662 | 0.09 |
ENST00000216252.3
|
PHF5A
|
PHD finger protein 5A |
| chr3_-_196295385 | 0.09 |
ENST00000425888.1
|
WDR53
|
WD repeat domain 53 |
| chr6_-_34664612 | 0.09 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr19_+_33182823 | 0.09 |
ENST00000397061.3
|
NUDT19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr2_-_202645612 | 0.09 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr14_+_103995503 | 0.09 |
ENST00000389749.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae) |
| chr1_-_11741155 | 0.09 |
ENST00000445656.1
ENST00000376669.5 ENST00000456915.1 ENST00000376692.4 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr19_-_5719860 | 0.09 |
ENST00000590729.1
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr16_-_790887 | 0.08 |
ENST00000540986.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr9_+_70856397 | 0.08 |
ENST00000360171.6
|
CBWD3
|
COBW domain containing 3 |
| chr17_-_58469329 | 0.08 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr2_-_240964716 | 0.08 |
ENST00000404554.1
ENST00000407129.3 ENST00000307300.4 ENST00000443626.1 ENST00000252711.2 |
NDUFA10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa |
| chr1_-_220219775 | 0.08 |
ENST00000609181.1
|
EPRS
|
glutamyl-prolyl-tRNA synthetase |
| chr8_-_37756972 | 0.08 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr19_-_54619006 | 0.08 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr2_+_234160217 | 0.08 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr8_+_25042192 | 0.08 |
ENST00000410074.1
|
DOCK5
|
dedicator of cytokinesis 5 |
| chr20_+_44420617 | 0.08 |
ENST00000449078.1
ENST00000456939.1 |
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr14_-_23388338 | 0.08 |
ENST00000555209.1
ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23
|
RNA binding motif protein 23 |
| chr7_-_1199781 | 0.08 |
ENST00000397083.1
ENST00000401903.1 ENST00000316495.3 |
ZFAND2A
|
zinc finger, AN1-type domain 2A |
| chr12_+_69633372 | 0.08 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr3_+_51428704 | 0.08 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr16_+_25123148 | 0.08 |
ENST00000570981.1
|
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr3_-_119182453 | 0.08 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr6_-_31939734 | 0.08 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr1_+_16085244 | 0.08 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr11_+_66610883 | 0.07 |
ENST00000309657.3
ENST00000524506.1 |
RCE1
|
Ras converting CAAX endopeptidase 1 |
| chr5_-_132112921 | 0.07 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr14_+_100150622 | 0.07 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr17_+_685513 | 0.07 |
ENST00000304478.4
|
RNMTL1
|
RNA methyltransferase like 1 |
| chr2_-_27579842 | 0.07 |
ENST00000423998.1
ENST00000264720.3 |
GTF3C2
|
general transcription factor IIIC, polypeptide 2, beta 110kDa |
| chr6_-_144416737 | 0.07 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr2_-_10830093 | 0.07 |
ENST00000381685.5
ENST00000345985.3 ENST00000542668.1 ENST00000538384.1 |
NOL10
|
nucleolar protein 10 |
| chr17_-_58469687 | 0.07 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr3_+_185303962 | 0.07 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr5_-_132112907 | 0.07 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr9_-_70490107 | 0.07 |
ENST00000377395.4
ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5
|
COBW domain containing 5 |
| chr5_-_72861484 | 0.07 |
ENST00000296785.3
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_-_5720248 | 0.07 |
ENST00000360614.3
|
LONP1
|
lon peptidase 1, mitochondrial |
| chr6_-_31926629 | 0.07 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr19_+_36120009 | 0.07 |
ENST00000589871.1
|
RBM42
|
RNA binding motif protein 42 |
| chr11_-_64570624 | 0.07 |
ENST00000439069.1
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr9_-_140082983 | 0.06 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr12_-_124118296 | 0.06 |
ENST00000424014.2
ENST00000537073.1 |
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr18_+_268148 | 0.06 |
ENST00000581677.1
|
RP11-705O1.8
|
RP11-705O1.8 |
| chr17_-_31404 | 0.06 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr16_+_88636789 | 0.06 |
ENST00000301011.5
ENST00000452588.2 |
ZC3H18
|
zinc finger CCCH-type containing 18 |
| chr8_-_126104055 | 0.06 |
ENST00000318410.7
|
KIAA0196
|
KIAA0196 |
| chr3_+_48488497 | 0.06 |
ENST00000412052.1
|
ATRIP
|
ATR interacting protein |
| chr9_-_86322831 | 0.06 |
ENST00000257468.7
|
UBQLN1
|
ubiquilin 1 |
| chr8_-_126103969 | 0.06 |
ENST00000517845.1
|
KIAA0196
|
KIAA0196 |
| chr4_-_54232144 | 0.06 |
ENST00000388940.4
ENST00000503450.1 ENST00000401642.3 |
SCFD2
|
sec1 family domain containing 2 |
| chr10_-_74114714 | 0.06 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr11_+_60609537 | 0.06 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 1.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.4 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.4 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.3 | GO:1904751 | positive regulation of telomeric DNA binding(GO:1904744) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.1 | 0.2 | GO:0045799 | negative regulation of nucleobase-containing compound transport(GO:0032240) positive regulation of chromatin assembly or disassembly(GO:0045799) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 | 0.6 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.1 | 0.2 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.2 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.0 | 0.4 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.2 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0098972 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0051664 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.1 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.1 | GO:0048320 | forebrain anterior/posterior pattern specification(GO:0021797) axial mesoderm formation(GO:0048320) |
| 0.0 | 0.0 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.1 | 0.3 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.4 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.4 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 0.3 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.2 | GO:0070362 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.2 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |