Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
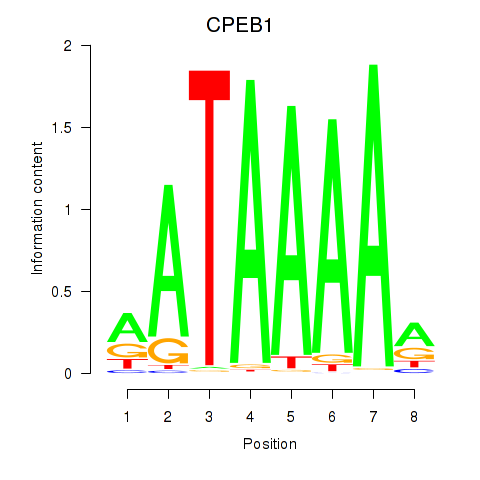
Results for CPEB1
Z-value: 1.02
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83316254_83316368 | 0.22 | 6.7e-01 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_159895275 | 1.56 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr8_-_25281747 | 0.88 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr4_-_74864386 | 0.86 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr10_-_106240032 | 0.76 |
ENST00000447860.1
|
RP11-127O4.3
|
RP11-127O4.3 |
| chr4_-_74904398 | 0.76 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr2_-_25451065 | 0.66 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr6_+_12290586 | 0.56 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr18_+_68002675 | 0.49 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr6_+_31105426 | 0.49 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr2_-_228244013 | 0.47 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr3_+_101546827 | 0.43 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_+_72058130 | 0.42 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_+_62284008 | 0.42 |
ENST00000544932.1
|
MTRNR2L9
|
MT-RNR2-like 9 (pseudogene) |
| chr12_-_14924056 | 0.38 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chr8_-_73793975 | 0.38 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr12_-_12714006 | 0.36 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_-_15038779 | 0.34 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr9_-_71869642 | 0.33 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr7_+_26591441 | 0.33 |
ENST00000420912.1
ENST00000457000.1 ENST00000430426.1 |
AC004947.2
|
AC004947.2 |
| chr15_-_79576156 | 0.33 |
ENST00000560452.1
ENST00000560872.1 ENST00000560732.1 ENST00000559979.1 ENST00000560533.1 ENST00000559225.1 |
RP11-17L5.4
|
RP11-17L5.4 |
| chr11_+_123301012 | 0.32 |
ENST00000533341.1
|
AP000783.1
|
Uncharacterized protein |
| chr18_+_29769978 | 0.32 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr4_+_74735102 | 0.31 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr17_+_7461580 | 0.30 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_+_94752349 | 0.30 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr21_-_35340759 | 0.29 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr17_-_2117600 | 0.29 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr3_-_141747439 | 0.29 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_-_69597828 | 0.28 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr20_-_10654639 | 0.26 |
ENST00000254958.5
|
JAG1
|
jagged 1 |
| chr11_-_102826434 | 0.26 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr2_+_152214098 | 0.26 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr17_+_7461613 | 0.26 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_-_61389240 | 0.26 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr4_+_76481258 | 0.26 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr11_-_102714534 | 0.25 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr3_+_99357319 | 0.25 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr12_-_12714025 | 0.25 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr8_+_30496078 | 0.24 |
ENST00000517349.1
|
SMIM18
|
small integral membrane protein 18 |
| chrX_-_100662881 | 0.24 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr7_-_83824449 | 0.24 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_+_66800928 | 0.23 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr5_+_36608280 | 0.23 |
ENST00000513646.1
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr17_+_7462103 | 0.23 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr8_+_77593474 | 0.23 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr11_-_102651343 | 0.22 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr5_+_32788945 | 0.22 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr12_-_56236734 | 0.22 |
ENST00000548629.1
|
MMP19
|
matrix metallopeptidase 19 |
| chr18_+_19749386 | 0.22 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr7_+_123488124 | 0.22 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr13_-_67802549 | 0.22 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr13_-_44735393 | 0.22 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr21_+_17792672 | 0.21 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr20_+_52105495 | 0.21 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr6_+_153071925 | 0.21 |
ENST00000367244.3
ENST00000367243.3 |
VIP
|
vasoactive intestinal peptide |
| chr16_+_2820912 | 0.20 |
ENST00000570539.1
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr17_-_39580775 | 0.20 |
ENST00000225550.3
|
KRT37
|
keratin 37 |
| chr17_+_79369249 | 0.20 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr19_-_58485895 | 0.20 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr1_+_152758690 | 0.20 |
ENST00000368771.1
ENST00000368770.3 |
LCE1E
|
late cornified envelope 1E |
| chr20_+_31823792 | 0.19 |
ENST00000375413.4
ENST00000354297.4 ENST00000375422.2 |
BPIFA1
|
BPI fold containing family A, member 1 |
| chr13_+_22245522 | 0.19 |
ENST00000382353.5
|
FGF9
|
fibroblast growth factor 9 |
| chr1_-_198906528 | 0.19 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr2_-_74726710 | 0.19 |
ENST00000377566.4
|
LBX2
|
ladybird homeobox 2 |
| chr18_+_61554932 | 0.18 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr22_+_29876197 | 0.18 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr5_-_115910630 | 0.18 |
ENST00000343348.6
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_+_190105909 | 0.17 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr3_+_195413160 | 0.17 |
ENST00000599448.1
|
LINC00969
|
long intergenic non-protein coding RNA 969 |
| chr14_+_79745682 | 0.17 |
ENST00000557594.1
|
NRXN3
|
neurexin 3 |
| chr11_-_57194817 | 0.17 |
ENST00000529748.1
ENST00000525474.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr10_+_114710516 | 0.16 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr9_-_16870704 | 0.16 |
ENST00000380672.4
ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2
|
basonuclin 2 |
| chr8_+_77596014 | 0.16 |
ENST00000523885.1
|
ZFHX4
|
zinc finger homeobox 4 |
| chr3_-_99569821 | 0.16 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr3_+_185046676 | 0.16 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr16_-_4164027 | 0.15 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr22_-_41258074 | 0.15 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr2_+_58655461 | 0.15 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr12_-_91546926 | 0.15 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_-_71353892 | 0.15 |
ENST00000484350.1
|
FOXP1
|
forkhead box P1 |
| chr5_-_88119580 | 0.15 |
ENST00000539796.1
|
MEF2C
|
myocyte enhancer factor 2C |
| chr22_+_39916558 | 0.15 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr12_+_8849773 | 0.15 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr9_-_133814455 | 0.15 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr12_-_52845910 | 0.15 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr6_+_30614886 | 0.15 |
ENST00000376471.4
|
C6orf136
|
chromosome 6 open reading frame 136 |
| chr17_-_47045949 | 0.15 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr17_+_32582293 | 0.14 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chrX_-_48931648 | 0.14 |
ENST00000376386.3
ENST00000376390.4 |
PRAF2
|
PRA1 domain family, member 2 |
| chr4_+_169575875 | 0.14 |
ENST00000503457.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr6_-_154751629 | 0.14 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr3_-_197300194 | 0.14 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr2_-_151344172 | 0.14 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr3_+_160394940 | 0.14 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr12_-_39734783 | 0.14 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr12_-_56236711 | 0.14 |
ENST00000409200.3
|
MMP19
|
matrix metallopeptidase 19 |
| chrM_+_14741 | 0.14 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chr12_-_123201337 | 0.14 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chrX_-_46187069 | 0.14 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr11_-_125773085 | 0.14 |
ENST00000227474.3
ENST00000534158.1 ENST00000529801.1 |
PUS3
|
pseudouridylate synthase 3 |
| chr7_+_28452130 | 0.14 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr8_+_77593448 | 0.13 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr2_+_40973618 | 0.13 |
ENST00000420187.1
|
AC007317.1
|
AC007317.1 |
| chr1_+_163039143 | 0.13 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr1_-_183560011 | 0.13 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr8_+_86351056 | 0.13 |
ENST00000285381.2
|
CA3
|
carbonic anhydrase III, muscle specific |
| chr3_-_99595037 | 0.13 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr3_+_44690211 | 0.13 |
ENST00000396056.2
ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35
|
zinc finger protein 35 |
| chr7_-_27219632 | 0.13 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr12_-_16760021 | 0.13 |
ENST00000540445.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr12_-_53228079 | 0.13 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr6_-_130543958 | 0.13 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr2_+_192542879 | 0.13 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr9_+_74920408 | 0.13 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chrX_-_19689106 | 0.12 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr13_-_31040060 | 0.12 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chr17_-_46806540 | 0.12 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr8_-_95220775 | 0.12 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr1_+_151739131 | 0.12 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr10_-_100027943 | 0.12 |
ENST00000260702.3
|
LOXL4
|
lysyl oxidase-like 4 |
| chr14_+_50291993 | 0.12 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr9_+_2717502 | 0.12 |
ENST00000382082.3
|
KCNV2
|
potassium channel, subfamily V, member 2 |
| chr15_+_48051920 | 0.12 |
ENST00000559196.1
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr1_-_32384693 | 0.12 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr1_+_209848749 | 0.11 |
ENST00000367029.4
|
G0S2
|
G0/G1switch 2 |
| chr10_+_114710425 | 0.11 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr17_+_7461849 | 0.11 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr14_-_75079026 | 0.11 |
ENST00000261978.4
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr17_+_7462031 | 0.11 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_41447609 | 0.11 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr12_-_116714564 | 0.11 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr1_+_241695424 | 0.11 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_30144432 | 0.11 |
ENST00000389048.3
|
ALK
|
anaplastic lymphoma receptor tyrosine kinase |
| chr2_+_163175394 | 0.11 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr12_+_54393880 | 0.11 |
ENST00000303450.4
|
HOXC9
|
homeobox C9 |
| chr3_+_51575596 | 0.11 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chrM_-_14670 | 0.11 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr3_+_57882061 | 0.11 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr10_+_115469134 | 0.11 |
ENST00000452490.2
|
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr9_+_82188077 | 0.11 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr17_-_10560619 | 0.11 |
ENST00000583535.1
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr2_+_192543153 | 0.11 |
ENST00000425611.2
|
NABP1
|
nucleic acid binding protein 1 |
| chrM_+_5824 | 0.11 |
ENST00000361624.2
|
MT-CO1
|
mitochondrially encoded cytochrome c oxidase I |
| chr7_-_111424462 | 0.11 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr3_-_73483055 | 0.11 |
ENST00000479530.1
|
PDZRN3
|
PDZ domain containing ring finger 3 |
| chr11_-_93583697 | 0.11 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr11_-_57194948 | 0.11 |
ENST00000533235.1
ENST00000526621.1 ENST00000352187.1 |
SLC43A3
|
solute carrier family 43, member 3 |
| chr5_+_36608422 | 0.10 |
ENST00000381918.3
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr7_-_27205136 | 0.10 |
ENST00000396345.1
ENST00000343483.6 |
HOXA9
|
homeobox A9 |
| chr12_-_52828147 | 0.10 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr15_-_60690163 | 0.10 |
ENST00000558998.1
ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2
|
annexin A2 |
| chrX_-_114252193 | 0.10 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr1_-_12679171 | 0.10 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr17_-_41739283 | 0.10 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr6_+_88054530 | 0.10 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr10_-_104262460 | 0.10 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr12_-_21927736 | 0.10 |
ENST00000240662.2
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr5_-_141704566 | 0.10 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr9_-_13165457 | 0.10 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr2_-_207023918 | 0.10 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr9_-_215744 | 0.10 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr3_-_15563229 | 0.10 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr4_+_117220016 | 0.10 |
ENST00000604093.1
|
MTRNR2L13
|
MT-RNR2-like 13 (pseudogene) |
| chr8_+_81397876 | 0.10 |
ENST00000430430.1
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr14_-_69261310 | 0.10 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr3_-_148939835 | 0.10 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_161677034 | 0.10 |
ENST00000349527.4
ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA
|
Fc receptor-like A |
| chr1_-_243349684 | 0.09 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr2_+_88047606 | 0.09 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr16_-_56459354 | 0.09 |
ENST00000290649.5
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr1_+_145883868 | 0.09 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr18_+_47088401 | 0.09 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chr3_-_114035026 | 0.09 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr6_+_161123270 | 0.09 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr1_+_169077133 | 0.09 |
ENST00000494797.1
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr3_-_10547192 | 0.09 |
ENST00000360273.2
ENST00000343816.4 |
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr12_+_85673868 | 0.09 |
ENST00000316824.3
|
ALX1
|
ALX homeobox 1 |
| chr1_+_154293584 | 0.09 |
ENST00000324978.3
ENST00000484864.1 |
AQP10
|
aquaporin 10 |
| chr8_+_85095769 | 0.09 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr6_-_130536774 | 0.09 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr3_-_10547333 | 0.09 |
ENST00000383800.4
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr7_-_18067478 | 0.09 |
ENST00000506618.2
|
PRPS1L1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr4_-_40632140 | 0.09 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr2_+_113763031 | 0.09 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr7_-_27219849 | 0.09 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chrX_+_133733457 | 0.09 |
ENST00000440614.1
|
RP11-308B5.2
|
RP11-308B5.2 |
| chr10_+_114710211 | 0.09 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr4_-_52904425 | 0.09 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr12_-_96794143 | 0.09 |
ENST00000543119.2
|
CDK17
|
cyclin-dependent kinase 17 |
| chr18_-_72920372 | 0.09 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr3_+_158787041 | 0.09 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr3_+_142342240 | 0.09 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr3_+_89156799 | 0.09 |
ENST00000452448.2
ENST00000494014.1 |
EPHA3
|
EPH receptor A3 |
| chr17_+_7461781 | 0.09 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_+_35160709 | 0.08 |
ENST00000415148.2
ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_241695670 | 0.08 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr7_-_111846435 | 0.08 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr6_+_30614779 | 0.08 |
ENST00000293604.6
ENST00000376473.5 |
C6orf136
|
chromosome 6 open reading frame 136 |
| chr1_+_107683436 | 0.08 |
ENST00000370068.1
|
NTNG1
|
netrin G1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 0.6 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.6 | GO:0060585 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 1.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 2.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.2 | GO:0051891 | positive regulation of cardioblast differentiation(GO:0051891) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.2 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.3 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:0044147 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.4 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.0 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.2 | GO:0051001 | glycoside catabolic process(GO:0016139) glycosylceramide catabolic process(GO:0046477) negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0061056 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:1902866 | regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0033007 | negative regulation of mast cell activation involved in immune response(GO:0033007) negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.0 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.1 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.0 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) |
| 0.0 | 0.1 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.0 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.4 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.9 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 2.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.9 | GO:0005179 | hormone activity(GO:0005179) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 2.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |