Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
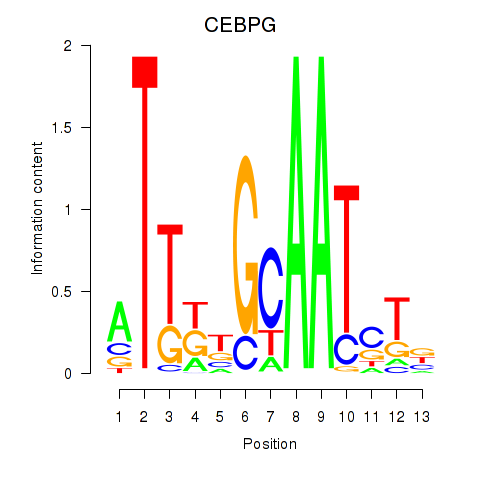
Results for CEBPG
Z-value: 1.97
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPG | hg19_v2_chr19_+_33865218_33865254 | -0.26 | 6.2e-01 | Click! |
Activity profile of CEBPG motif
Sorted Z-values of CEBPG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22766766 | 5.39 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr11_-_18270182 | 4.42 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr11_+_18287801 | 4.33 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_+_18287721 | 4.31 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr1_-_47655686 | 3.32 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr6_+_31895467 | 3.09 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr1_-_153321301 | 2.84 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr6_+_31895480 | 2.79 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr11_-_18258342 | 2.74 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr2_-_113594279 | 1.90 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr19_-_6720686 | 1.87 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr12_+_72058130 | 1.62 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr6_-_133055815 | 1.59 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr6_-_133055896 | 1.54 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr19_-_6670128 | 1.53 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr9_+_130911723 | 1.40 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr3_-_4793274 | 1.24 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr11_-_102668879 | 1.16 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr19_-_4540486 | 1.09 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr9_+_130911770 | 1.04 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr22_-_30901637 | 0.99 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr5_-_121413974 | 0.78 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr14_-_23292596 | 0.74 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr14_+_39703112 | 0.74 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr17_-_79895097 | 0.66 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr10_+_26986582 | 0.65 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr12_+_27849378 | 0.60 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr7_-_112635675 | 0.56 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr2_-_191885686 | 0.54 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr17_+_812872 | 0.54 |
ENST00000576252.1
|
RP11-676J12.7
|
Uncharacterized protein |
| chr17_-_79895154 | 0.54 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr22_-_42765174 | 0.51 |
ENST00000432473.1
ENST00000412060.1 ENST00000424852.1 |
Z83851.1
|
Z83851.1 |
| chr19_+_49497121 | 0.48 |
ENST00000413176.2
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr12_-_95510743 | 0.48 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr11_-_33913708 | 0.48 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr19_+_49496705 | 0.47 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_+_49496782 | 0.47 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr1_+_165864800 | 0.46 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr17_+_4699439 | 0.45 |
ENST00000270586.3
|
PSMB6
|
proteasome (prosome, macropain) subunit, beta type, 6 |
| chr19_-_49496557 | 0.45 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr3_-_148939598 | 0.44 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr6_+_31895254 | 0.44 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr6_+_37897735 | 0.43 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr21_+_33671264 | 0.43 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr2_-_216300784 | 0.42 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr21_+_33671160 | 0.41 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr21_+_34775181 | 0.41 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr20_+_361261 | 0.41 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr14_-_70263979 | 0.40 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr20_-_48782639 | 0.39 |
ENST00000435301.2
|
RP11-112L6.3
|
RP11-112L6.3 |
| chr3_+_157154578 | 0.39 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr7_-_14026063 | 0.36 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr21_+_43619796 | 0.36 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr1_-_243326612 | 0.35 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr17_-_39769005 | 0.33 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chrX_-_100129128 | 0.33 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr21_-_32119551 | 0.33 |
ENST00000333892.2
|
KRTAP21-2
|
keratin associated protein 21-2 |
| chr2_-_175711978 | 0.31 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chrX_+_47082408 | 0.30 |
ENST00000518022.1
ENST00000276052.6 |
CDK16
|
cyclin-dependent kinase 16 |
| chr5_+_125759140 | 0.30 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr12_+_53846612 | 0.29 |
ENST00000551104.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr21_-_35899113 | 0.28 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr15_+_67813406 | 0.28 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr7_-_107968999 | 0.27 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr12_-_57039739 | 0.26 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr6_+_31895287 | 0.25 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr9_+_117085336 | 0.24 |
ENST00000259396.8
ENST00000538816.1 |
ORM1
|
orosomucoid 1 |
| chr17_+_7477040 | 0.24 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr19_+_45394477 | 0.24 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr3_-_120400960 | 0.23 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr1_+_165864821 | 0.23 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr5_+_125758813 | 0.23 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chrX_+_46937745 | 0.23 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chr3_-_157221128 | 0.22 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr7_-_83824449 | 0.22 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr14_+_45366472 | 0.21 |
ENST00000325192.3
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr5_+_49962772 | 0.21 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr7_+_137761199 | 0.21 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr4_-_119274121 | 0.20 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr10_+_18629628 | 0.18 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr12_-_71551652 | 0.17 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr5_+_36152091 | 0.17 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr9_+_130860810 | 0.17 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr4_+_74269956 | 0.17 |
ENST00000295897.4
ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB
|
albumin |
| chr12_-_10605929 | 0.16 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr22_-_31364187 | 0.16 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr20_+_361890 | 0.16 |
ENST00000449710.1
ENST00000422053.2 |
TRIB3
|
tribbles pseudokinase 3 |
| chr7_-_8276508 | 0.16 |
ENST00000401396.1
ENST00000317367.5 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chrX_-_73512411 | 0.15 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr3_+_186435065 | 0.15 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr1_-_168698433 | 0.15 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr11_-_72504681 | 0.15 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr1_+_111770294 | 0.15 |
ENST00000474304.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr19_-_36822595 | 0.15 |
ENST00000585356.1
ENST00000438368.2 ENST00000590622.1 |
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr5_-_146258205 | 0.15 |
ENST00000394413.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_-_73512358 | 0.14 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr5_-_146258291 | 0.14 |
ENST00000394411.4
ENST00000453001.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr11_+_60048053 | 0.13 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr17_-_76124711 | 0.13 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr1_+_158901329 | 0.12 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr18_+_33767473 | 0.12 |
ENST00000261326.5
|
MOCOS
|
molybdenum cofactor sulfurase |
| chr1_+_196743912 | 0.12 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr1_-_161277210 | 0.12 |
ENST00000491222.2
|
MPZ
|
myelin protein zero |
| chr19_-_39805976 | 0.12 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr4_-_111120132 | 0.12 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr10_-_14596140 | 0.12 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr10_+_17272608 | 0.11 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr2_-_112614424 | 0.11 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr1_+_75594119 | 0.11 |
ENST00000294638.5
|
LHX8
|
LIM homeobox 8 |
| chr4_+_185909970 | 0.11 |
ENST00000505053.1
|
RP11-386B13.4
|
RP11-386B13.4 |
| chr19_-_36822551 | 0.10 |
ENST00000591372.1
|
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr1_+_111770232 | 0.10 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chrX_-_153775047 | 0.09 |
ENST00000433845.1
ENST00000439227.1 |
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr11_-_40315640 | 0.09 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr21_-_37852359 | 0.09 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr12_-_71551868 | 0.09 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr1_-_114430169 | 0.09 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr7_-_14026123 | 0.09 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr1_+_172389821 | 0.08 |
ENST00000367727.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr5_+_125758865 | 0.08 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_72504637 | 0.08 |
ENST00000536377.1
ENST00000359373.5 |
STARD10
ARAP1
|
StAR-related lipid transfer (START) domain containing 10 ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr17_+_43238061 | 0.08 |
ENST00000307275.3
ENST00000585340.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr11_+_60048129 | 0.08 |
ENST00000355131.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr3_-_148939835 | 0.08 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_+_53835508 | 0.08 |
ENST00000551003.1
ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13
PCBP2
|
proline rich 13 poly(rC) binding protein 2 |
| chr3_-_190580404 | 0.08 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr12_-_57522813 | 0.07 |
ENST00000556155.1
|
STAT6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr12_+_53846594 | 0.07 |
ENST00000550192.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr4_+_184020398 | 0.07 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr16_+_72088376 | 0.07 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr17_+_2699697 | 0.06 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr7_-_121944491 | 0.06 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr13_-_72441315 | 0.06 |
ENST00000305425.4
ENST00000313174.7 ENST00000354591.4 |
DACH1
|
dachshund homolog 1 (Drosophila) |
| chr4_+_175839506 | 0.06 |
ENST00000505141.1
ENST00000359240.3 ENST00000445694.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr7_-_144435985 | 0.06 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr1_-_179112173 | 0.05 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr22_-_50523760 | 0.05 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr2_+_218994002 | 0.05 |
ENST00000428565.1
|
CXCR2
|
chemokine (C-X-C motif) receptor 2 |
| chrX_-_73511908 | 0.05 |
ENST00000455395.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr12_+_53835383 | 0.05 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr4_+_175839551 | 0.04 |
ENST00000404450.4
ENST00000514159.1 |
ADAM29
|
ADAM metallopeptidase domain 29 |
| chr3_-_49726486 | 0.04 |
ENST00000449682.2
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr4_-_111119804 | 0.04 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr20_-_60294804 | 0.04 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr9_+_115913222 | 0.04 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr19_+_2476116 | 0.03 |
ENST00000215631.4
ENST00000587345.1 |
GADD45B
|
growth arrest and DNA-damage-inducible, beta |
| chr6_-_27799305 | 0.03 |
ENST00000357549.2
|
HIST1H4K
|
histone cluster 1, H4k |
| chr17_+_57287228 | 0.03 |
ENST00000578922.1
ENST00000300917.5 |
SMG8
|
SMG8 nonsense mediated mRNA decay factor |
| chr2_+_47454054 | 0.03 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr2_+_226265364 | 0.03 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr19_-_40971667 | 0.02 |
ENST00000263368.4
|
BLVRB
|
biliverdin reductase B (flavin reductase (NADPH)) |
| chr3_+_73110810 | 0.02 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr12_-_54758251 | 0.02 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chr2_-_183291741 | 0.02 |
ENST00000351439.5
ENST00000409365.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr19_-_1155118 | 0.02 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr13_-_36788718 | 0.02 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr3_+_186330712 | 0.02 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr5_-_135701164 | 0.02 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr19_+_6887571 | 0.01 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr1_-_179112189 | 0.01 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr11_-_124806297 | 0.01 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr22_-_30867973 | 0.01 |
ENST00000402286.1
ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3
|
SEC14-like 3 (S. cerevisiae) |
| chr2_+_113825547 | 0.01 |
ENST00000341010.2
ENST00000337569.3 |
IL1F10
|
interleukin 1 family, member 10 (theta) |
| chr16_-_57219721 | 0.01 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr1_+_111770278 | 0.01 |
ENST00000369748.4
|
CHI3L2
|
chitinase 3-like 2 |
| chr4_+_71600144 | 0.01 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_-_1711508 | 0.01 |
ENST00000378625.1
|
NADK
|
NAD kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.9 | 2.8 | GO:0051714 | positive regulation of cytolysis in other organism(GO:0051714) |
| 0.6 | 1.9 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.5 | 2.4 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 5.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 1.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 1.4 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.9 | GO:0031622 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.2 | 14.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 3.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 0.7 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.2 | 1.2 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.4 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.2 | GO:0044179 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 0.3 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of sperm motility(GO:1901318) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 1.9 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.6 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.3 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.8 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.4 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.4 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 15.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 8.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 3.5 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.4 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.6 | 3.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 15.8 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.3 | 2.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 0.8 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.2 | 1.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 6.3 | GO:0001848 | complement binding(GO:0001848) |
| 0.2 | 1.4 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 1.9 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.8 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 0.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.5 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.6 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 7.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.2 | 5.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 8.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 3.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |