Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
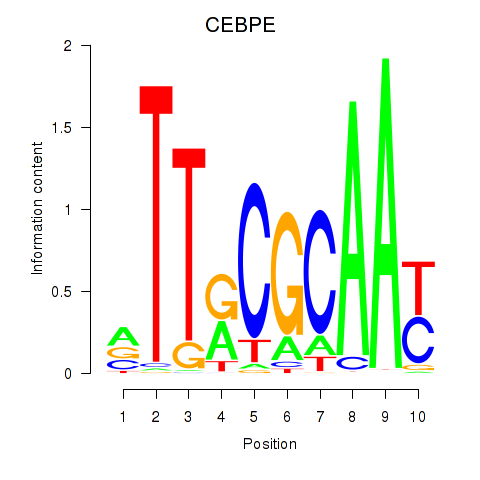
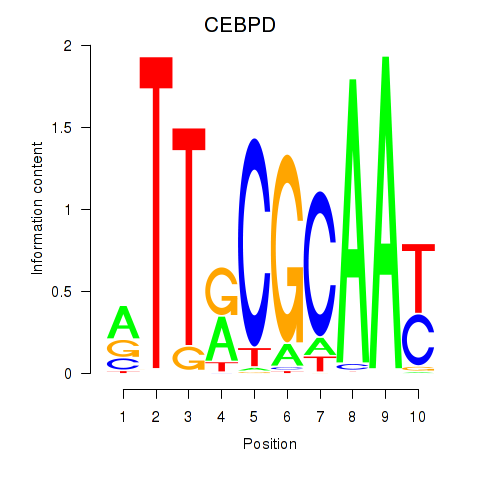
Results for CEBPE_CEBPD
Z-value: 1.39
Transcription factors associated with CEBPE_CEBPD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPE
|
ENSG00000092067.5 | CCAAT enhancer binding protein epsilon |
|
CEBPD
|
ENSG00000221869.4 | CCAAT enhancer binding protein delta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPD | hg19_v2_chr8_-_48651648_48651648 | -0.38 | 4.6e-01 | Click! |
Activity profile of CEBPE_CEBPD motif
Sorted Z-values of CEBPE_CEBPD motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22766766 | 4.53 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr11_-_18270182 | 3.75 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr11_+_18287801 | 3.43 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_+_18287721 | 3.41 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr6_-_133055815 | 3.33 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr6_-_133055896 | 3.26 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr6_+_31895467 | 2.27 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_+_31895480 | 2.11 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr19_-_6720686 | 2.04 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr11_+_93754513 | 2.03 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr2_-_113594279 | 1.54 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr12_+_7167980 | 0.98 |
ENST00000360817.5
ENST00000402681.3 |
C1S
|
complement component 1, s subcomponent |
| chr8_-_11324273 | 0.96 |
ENST00000284486.4
|
FAM167A
|
family with sequence similarity 167, member A |
| chr11_-_18258342 | 0.96 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr12_+_102271129 | 0.93 |
ENST00000258534.8
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr3_-_4793274 | 0.86 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr12_-_123201337 | 0.83 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr10_+_30723045 | 0.77 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr1_+_144339738 | 0.77 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr10_+_30723105 | 0.76 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr3_-_158450231 | 0.75 |
ENST00000479756.1
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr12_+_72058130 | 0.73 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr2_-_31637560 | 0.73 |
ENST00000379416.3
|
XDH
|
xanthine dehydrogenase |
| chr5_-_147286065 | 0.71 |
ENST00000318315.4
ENST00000515291.1 |
C5orf46
|
chromosome 5 open reading frame 46 |
| chr8_+_124194875 | 0.68 |
ENST00000522648.1
ENST00000276699.6 |
FAM83A
|
family with sequence similarity 83, member A |
| chr12_+_102271436 | 0.66 |
ENST00000544152.1
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_-_186649543 | 0.63 |
ENST00000367468.5
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_+_169079823 | 0.59 |
ENST00000367813.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr15_+_45926919 | 0.59 |
ENST00000561735.1
ENST00000260324.7 |
SQRDL
|
sulfide quinone reductase-like (yeast) |
| chr6_-_31940065 | 0.58 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr19_-_6459746 | 0.56 |
ENST00000301454.4
ENST00000334510.5 |
SLC25A23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr8_+_24151553 | 0.53 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr21_-_32119551 | 0.53 |
ENST00000333892.2
|
KRTAP21-2
|
keratin associated protein 21-2 |
| chr19_-_6670128 | 0.52 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr14_-_38028689 | 0.51 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr7_+_90032821 | 0.51 |
ENST00000427904.1
|
CLDN12
|
claudin 12 |
| chr20_-_43883197 | 0.50 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr21_+_33671264 | 0.48 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr5_-_121413974 | 0.47 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr14_+_20937538 | 0.47 |
ENST00000361505.5
ENST00000553591.1 |
PNP
|
purine nucleoside phosphorylase |
| chr14_+_102276192 | 0.43 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr5_-_138718973 | 0.43 |
ENST00000353963.3
ENST00000348729.3 |
SLC23A1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr12_-_123187890 | 0.42 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr1_+_209941942 | 0.42 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr1_-_45956868 | 0.41 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr20_-_17662878 | 0.41 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr14_-_64971288 | 0.41 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr3_-_158450475 | 0.40 |
ENST00000237696.5
|
RARRES1
|
retinoic acid receptor responder (tazarotene induced) 1 |
| chr17_+_7462103 | 0.39 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr14_+_64971438 | 0.39 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr17_-_79895097 | 0.39 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr8_+_124194752 | 0.37 |
ENST00000318462.6
|
FAM83A
|
family with sequence similarity 83, member A |
| chr1_-_156721502 | 0.37 |
ENST00000357325.5
|
HDGF
|
hepatoma-derived growth factor |
| chr16_-_84651673 | 0.36 |
ENST00000262428.4
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr14_-_70263979 | 0.36 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr3_-_148939598 | 0.36 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_-_84651647 | 0.36 |
ENST00000564057.1
|
COTL1
|
coactosin-like 1 (Dictyostelium) |
| chr19_-_41945804 | 0.35 |
ENST00000221943.9
ENST00000597457.1 ENST00000589970.1 ENST00000595425.1 ENST00000438807.3 ENST00000589102.1 ENST00000592922.2 |
ATP5SL
|
ATP5S-like |
| chr2_+_113885138 | 0.34 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr12_+_66582919 | 0.34 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr2_-_191878162 | 0.34 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr4_-_170192000 | 0.34 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr2_-_175711978 | 0.32 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr21_+_33671160 | 0.31 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr6_+_31895254 | 0.31 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr17_-_79895154 | 0.31 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr14_-_23292596 | 0.31 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_+_157154578 | 0.30 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr2_-_216300784 | 0.30 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr14_-_45722360 | 0.29 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr10_+_30723533 | 0.29 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr3_+_52279737 | 0.28 |
ENST00000457351.2
|
PPM1M
|
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr7_-_130597935 | 0.28 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr10_+_60028818 | 0.28 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr20_+_58630972 | 0.27 |
ENST00000313426.1
|
C20orf197
|
chromosome 20 open reading frame 197 |
| chr6_+_31515337 | 0.24 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr20_-_43743790 | 0.24 |
ENST00000307971.4
ENST00000372789.4 |
WFDC5
|
WAP four-disulfide core domain 5 |
| chr20_+_33759854 | 0.23 |
ENST00000216968.4
|
PROCR
|
protein C receptor, endothelial |
| chr1_-_47697387 | 0.23 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr17_+_7477040 | 0.23 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr8_+_24151620 | 0.22 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr6_-_24489842 | 0.21 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr2_-_191878874 | 0.20 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr11_-_47447970 | 0.20 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr6_+_101847105 | 0.20 |
ENST00000369137.3
ENST00000318991.6 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chrX_-_40506766 | 0.20 |
ENST00000378421.1
ENST00000440784.2 ENST00000327877.5 ENST00000378426.1 ENST00000378418.2 |
CXorf38
|
chromosome X open reading frame 38 |
| chr7_-_83824449 | 0.20 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_79817091 | 0.20 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr14_+_102276132 | 0.19 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_99594948 | 0.19 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr2_-_191878681 | 0.19 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr5_-_169407744 | 0.19 |
ENST00000377365.3
|
FAM196B
|
family with sequence similarity 196, member B |
| chr19_+_45174724 | 0.19 |
ENST00000358777.4
|
CEACAM19
|
carcinoembryonic antigen-related cell adhesion molecule 19 |
| chr10_+_14880157 | 0.19 |
ENST00000378372.3
|
HSPA14
|
heat shock 70kDa protein 14 |
| chrX_-_73512411 | 0.18 |
ENST00000602576.1
ENST00000429124.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr5_-_146435694 | 0.18 |
ENST00000356826.3
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr6_+_1389989 | 0.18 |
ENST00000259806.1
|
FOXF2
|
forkhead box F2 |
| chr5_+_49962772 | 0.18 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr15_+_63050785 | 0.18 |
ENST00000472902.1
|
TLN2
|
talin 2 |
| chr11_-_72145426 | 0.18 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr2_-_191885686 | 0.18 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr4_+_55095264 | 0.18 |
ENST00000257290.5
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr1_+_162531294 | 0.17 |
ENST00000367926.4
ENST00000271469.3 |
UAP1
|
UDP-N-acteylglucosamine pyrophosphorylase 1 |
| chr3_+_11313995 | 0.17 |
ENST00000451513.1
ENST00000435760.1 ENST00000451830.1 |
ATG7
|
autophagy related 7 |
| chr8_-_6735451 | 0.16 |
ENST00000297439.3
|
DEFB1
|
defensin, beta 1 |
| chr12_+_48592134 | 0.16 |
ENST00000595310.1
|
DKFZP779L1853
|
DKFZP779L1853 |
| chr1_-_161087802 | 0.16 |
ENST00000368010.3
|
PFDN2
|
prefoldin subunit 2 |
| chr6_+_31895287 | 0.16 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr22_-_20731541 | 0.16 |
ENST00000292729.8
|
USP41
|
ubiquitin specific peptidase 41 |
| chr12_-_91573249 | 0.16 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr1_+_165797024 | 0.16 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr10_-_128994422 | 0.16 |
ENST00000522781.1
|
FAM196A
|
family with sequence similarity 196, member A |
| chr19_+_39903185 | 0.15 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr1_+_209941827 | 0.15 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr3_+_11314072 | 0.15 |
ENST00000444619.1
|
ATG7
|
autophagy related 7 |
| chr15_+_67418047 | 0.15 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr7_-_87936195 | 0.15 |
ENST00000414498.1
ENST00000301959.5 ENST00000380079.4 |
STEAP4
|
STEAP family member 4 |
| chr3_+_136649311 | 0.14 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr5_-_137090028 | 0.14 |
ENST00000314940.4
|
HNRNPA0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr6_-_112194484 | 0.13 |
ENST00000518295.1
ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr11_-_47447767 | 0.13 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr11_-_8739383 | 0.13 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr12_+_106994905 | 0.13 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr2_+_219246746 | 0.13 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr13_-_21634421 | 0.13 |
ENST00000542899.1
|
LATS2
|
large tumor suppressor kinase 2 |
| chr1_+_196743912 | 0.12 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr3_+_11314099 | 0.12 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr15_+_39873268 | 0.12 |
ENST00000397591.2
ENST00000260356.5 |
THBS1
|
thrombospondin 1 |
| chrX_-_15620192 | 0.12 |
ENST00000427411.1
|
ACE2
|
angiotensin I converting enzyme 2 |
| chrX_-_73512358 | 0.12 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr17_+_7461849 | 0.11 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr21_+_43619796 | 0.11 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr3_-_99595037 | 0.11 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr7_+_150065278 | 0.11 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr17_+_27181956 | 0.11 |
ENST00000254928.5
ENST00000580917.2 |
ERAL1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr16_+_72088376 | 0.11 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr1_+_10093188 | 0.10 |
ENST00000377153.1
|
UBE4B
|
ubiquitination factor E4B |
| chr21_+_27011584 | 0.10 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr9_+_130860810 | 0.10 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr10_-_75226166 | 0.10 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr16_-_70323422 | 0.10 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr20_+_4667094 | 0.10 |
ENST00000424424.1
ENST00000457586.1 |
PRNP
|
prion protein |
| chr5_-_58882219 | 0.10 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_+_12392971 | 0.09 |
ENST00000287820.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr4_+_25657444 | 0.09 |
ENST00000504570.1
ENST00000382051.3 |
SLC34A2
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 2 |
| chr1_+_222910625 | 0.09 |
ENST00000360827.2
|
FAM177B
|
family with sequence similarity 177, member B |
| chr2_+_65663812 | 0.09 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chrX_-_119445306 | 0.09 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr4_-_155511887 | 0.09 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr6_+_4776580 | 0.09 |
ENST00000397588.3
|
CDYL
|
chromodomain protein, Y-like |
| chrM_+_7586 | 0.09 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr20_-_1600642 | 0.08 |
ENST00000381603.3
ENST00000381605.4 ENST00000279477.7 ENST00000568365.1 ENST00000564763.1 |
SIRPB1
RP4-576H24.4
|
signal-regulatory protein beta 1 Uncharacterized protein |
| chr20_-_60294804 | 0.08 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr2_+_234296792 | 0.08 |
ENST00000409813.3
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr12_+_110011571 | 0.08 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr17_+_7348658 | 0.08 |
ENST00000570557.1
ENST00000536404.2 ENST00000576360.1 |
CHRNB1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr13_-_24471641 | 0.08 |
ENST00000382145.1
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr4_-_186732241 | 0.08 |
ENST00000421639.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chrX_+_49126294 | 0.08 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr2_-_157186630 | 0.08 |
ENST00000406048.2
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_-_121944491 | 0.08 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chrX_+_46937745 | 0.07 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chr21_+_17792672 | 0.07 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_7461781 | 0.07 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr21_-_46334186 | 0.07 |
ENST00000522931.1
|
ITGB2
|
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr7_-_73153122 | 0.07 |
ENST00000458339.1
|
ABHD11
|
abhydrolase domain containing 11 |
| chr4_+_55095428 | 0.07 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr10_-_97416400 | 0.07 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr15_+_58430567 | 0.07 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chrX_-_102941596 | 0.07 |
ENST00000441076.2
ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2
|
mortality factor 4 like 2 |
| chr3_-_190580404 | 0.07 |
ENST00000442080.1
|
GMNC
|
geminin coiled-coil domain containing |
| chr12_+_110940111 | 0.07 |
ENST00000409778.3
|
RAD9B
|
RAD9 homolog B (S. pombe) |
| chr14_-_103523745 | 0.07 |
ENST00000361246.2
|
CDC42BPB
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr1_-_155829191 | 0.06 |
ENST00000437809.1
|
GON4L
|
gon-4-like (C. elegans) |
| chr11_-_40315640 | 0.06 |
ENST00000278198.2
|
LRRC4C
|
leucine rich repeat containing 4C |
| chr20_+_56136136 | 0.06 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chrX_-_135962923 | 0.06 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr20_-_33732952 | 0.06 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_-_99381660 | 0.06 |
ENST00000375240.3
ENST00000463569.1 |
CDC14B
|
cell division cycle 14B |
| chr15_+_58430368 | 0.06 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr2_+_33683109 | 0.06 |
ENST00000437184.1
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr7_+_137761199 | 0.06 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr1_-_92951607 | 0.05 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr2_+_68694678 | 0.05 |
ENST00000303795.4
|
APLF
|
aprataxin and PNKP like factor |
| chr2_-_46769694 | 0.05 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr10_+_14880364 | 0.05 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr3_+_121902511 | 0.05 |
ENST00000490131.1
|
CASR
|
calcium-sensing receptor |
| chr17_+_7462031 | 0.05 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr7_-_36764004 | 0.05 |
ENST00000431169.1
|
AOAH
|
acyloxyacyl hydrolase (neutrophil) |
| chr3_-_120400960 | 0.05 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr19_+_48949030 | 0.05 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr1_+_161087873 | 0.05 |
ENST00000368009.2
ENST00000368007.4 ENST00000368008.1 ENST00000392190.5 |
NIT1
|
nitrilase 1 |
| chr1_+_111770294 | 0.04 |
ENST00000474304.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr1_-_168698433 | 0.04 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chrX_-_119445263 | 0.04 |
ENST00000309720.5
|
TMEM255A
|
transmembrane protein 255A |
| chr16_+_3507985 | 0.04 |
ENST00000421765.3
ENST00000360862.5 ENST00000414063.2 ENST00000610180.1 ENST00000608993.1 |
NAA60
NAA60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit N-alpha-acetyltransferase 60 |
| chr10_-_17659234 | 0.04 |
ENST00000466335.1
|
PTPLA
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr8_+_77318769 | 0.04 |
ENST00000518732.1
|
RP11-706J10.1
|
long intergenic non-protein coding RNA 1111 |
| chr5_+_140227357 | 0.04 |
ENST00000378122.3
|
PCDHA9
|
protocadherin alpha 9 |
| chr15_-_53002007 | 0.04 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPE_CEBPD
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.7 | 2.0 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.4 | 6.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.3 | 2.2 | GO:0031620 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.2 | 4.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 0.5 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.2 | 0.9 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.2 | 0.7 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.2 | 6.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.4 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 0.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 | 0.8 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.3 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.1 | 0.6 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.1 | 0.2 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.1 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 2.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.2 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.6 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0010983 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.3 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 4.9 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.2 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0021824 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) neural crest cell migration involved in sympathetic nervous system development(GO:1903045) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0001828 | inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.6 | GO:0071027 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of sperm motility(GO:1901318) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.7 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0071918 | pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.5 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.2 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0051495 | positive regulation of cytoskeleton organization(GO:0051495) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 6.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 6.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 4.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.3 | 0.8 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.2 | 11.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 2.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.6 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.2 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.4 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.1 | 4.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.6 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.7 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.1 | 0.7 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.1 | 1.6 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) collagen V binding(GO:0070052) |
| 0.0 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0015265 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 4.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 6.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 4.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.8 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 6.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 4.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |