Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
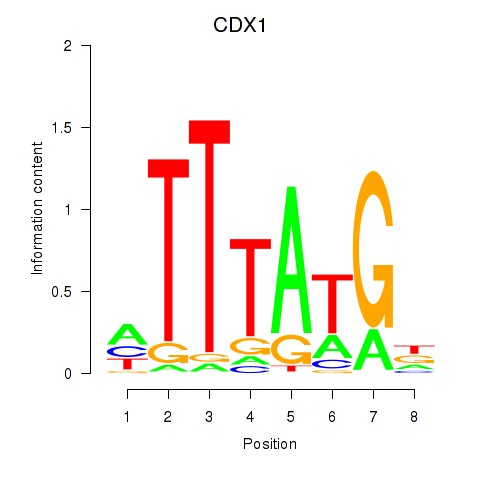
Results for CDX1
Z-value: 0.61
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | caudal type homeobox 1 |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_35013217 | 0.71 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr8_+_103876528 | 0.61 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chr6_+_63921351 | 0.57 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr1_+_212475148 | 0.34 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr8_-_42358742 | 0.32 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr14_-_54955721 | 0.32 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr18_-_59415987 | 0.30 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr12_-_76461249 | 0.29 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr5_+_118691706 | 0.28 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_-_57229155 | 0.28 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr18_+_61575200 | 0.28 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr10_-_18948208 | 0.28 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr5_+_115177178 | 0.27 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr8_-_125551278 | 0.27 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr16_-_18887627 | 0.27 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chrX_-_119693745 | 0.26 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr14_-_60952739 | 0.25 |
ENST00000555476.1
ENST00000321731.3 |
C14orf39
|
chromosome 14 open reading frame 39 |
| chr8_-_123706338 | 0.25 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr3_-_96337000 | 0.24 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr3_+_182983126 | 0.24 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr7_+_79765071 | 0.23 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr21_+_17214724 | 0.22 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr1_-_193075180 | 0.21 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr1_+_152730499 | 0.21 |
ENST00000368773.1
|
KPRP
|
keratinocyte proline-rich protein |
| chr4_-_175205407 | 0.20 |
ENST00000393674.2
|
FBXO8
|
F-box protein 8 |
| chr17_-_39553844 | 0.20 |
ENST00000251645.2
|
KRT31
|
keratin 31 |
| chr7_-_54826920 | 0.19 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr5_+_102200948 | 0.19 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr14_+_37641012 | 0.19 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr21_-_35773370 | 0.19 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr12_-_91573132 | 0.19 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr17_+_61151306 | 0.19 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr7_-_33080506 | 0.18 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr14_+_61201445 | 0.18 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr8_+_94752349 | 0.18 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr21_+_25801041 | 0.17 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr8_+_7705398 | 0.17 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr21_-_43735628 | 0.16 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr7_+_105172532 | 0.16 |
ENST00000257700.2
|
RINT1
|
RAD50 interactor 1 |
| chr18_+_47901408 | 0.16 |
ENST00000398452.2
ENST00000417656.2 ENST00000488454.1 ENST00000494518.1 |
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr15_+_49715449 | 0.16 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr11_+_112046190 | 0.16 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr18_-_68004529 | 0.16 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr8_-_7320974 | 0.16 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr11_-_85780853 | 0.16 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr4_+_71091786 | 0.16 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr13_+_28519343 | 0.15 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr9_-_69229650 | 0.15 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr3_-_196987309 | 0.15 |
ENST00000453607.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr19_+_52076425 | 0.15 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr1_-_68299130 | 0.15 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr4_+_154622652 | 0.15 |
ENST00000260010.6
|
TLR2
|
toll-like receptor 2 |
| chr10_-_36813162 | 0.14 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr1_-_111970353 | 0.14 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr3_-_149095652 | 0.14 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr3_-_196911002 | 0.14 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr5_-_95550754 | 0.14 |
ENST00000502437.1
|
RP11-254I22.3
|
RP11-254I22.3 |
| chr8_-_112248400 | 0.14 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr18_+_55712915 | 0.14 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_54411240 | 0.14 |
ENST00000371378.2
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr12_+_97306295 | 0.14 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr17_-_59668550 | 0.14 |
ENST00000521764.1
|
NACA2
|
nascent polypeptide-associated complex alpha subunit 2 |
| chr3_+_182983090 | 0.14 |
ENST00000465010.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr11_-_5537920 | 0.13 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr3_+_44916098 | 0.13 |
ENST00000296125.4
|
TGM4
|
transglutaminase 4 |
| chr9_-_3469181 | 0.13 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr2_+_89901292 | 0.13 |
ENST00000448155.2
|
IGKV1D-39
|
immunoglobulin kappa variable 1D-39 |
| chr6_+_116850174 | 0.13 |
ENST00000416171.2
ENST00000368597.2 ENST00000452373.1 ENST00000405399.1 |
FAM26D
|
family with sequence similarity 26, member D |
| chr2_+_242289502 | 0.13 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr1_+_84629976 | 0.13 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_+_28410128 | 0.13 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr17_+_65713925 | 0.13 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr1_+_84630574 | 0.12 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr14_+_58797974 | 0.12 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr7_-_92777606 | 0.12 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chrX_-_45629661 | 0.12 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr5_+_138210919 | 0.12 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr2_-_89619904 | 0.12 |
ENST00000498574.1
|
IGKV1-39
|
immunoglobulin kappa variable 1-39 (gene/pseudogene) |
| chr5_+_118691008 | 0.12 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_15986364 | 0.12 |
ENST00000345034.1
|
RSC1A1
|
regulatory solute carrier protein, family 1, member 1 |
| chr7_+_120629653 | 0.12 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr14_-_67826538 | 0.12 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr4_-_100065440 | 0.12 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr16_+_53241854 | 0.12 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr22_+_21128167 | 0.12 |
ENST00000215727.5
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr3_-_33686925 | 0.12 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr11_+_114310164 | 0.12 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr18_+_61445007 | 0.12 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr10_+_118187379 | 0.11 |
ENST00000369230.3
|
PNLIPRP3
|
pancreatic lipase-related protein 3 |
| chr1_+_84630053 | 0.11 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr12_+_28605426 | 0.11 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_935491 | 0.11 |
ENST00000304952.6
|
HES4
|
hes family bHLH transcription factor 4 |
| chr11_+_28129795 | 0.11 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr4_+_86748898 | 0.11 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr14_-_31926623 | 0.11 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr8_-_103876383 | 0.10 |
ENST00000347770.4
|
AZIN1
|
antizyme inhibitor 1 |
| chr2_+_172309634 | 0.10 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr17_-_55911970 | 0.10 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr15_+_45879964 | 0.10 |
ENST00000565409.1
ENST00000564765.1 |
BLOC1S6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr9_-_21482312 | 0.10 |
ENST00000448696.3
|
IFNE
|
interferon, epsilon |
| chr5_+_140602904 | 0.10 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr3_+_167582561 | 0.10 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr3_-_168865522 | 0.10 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr1_+_200993071 | 0.10 |
ENST00000446333.1
ENST00000458003.1 |
RP11-168O16.1
|
RP11-168O16.1 |
| chr12_+_50690489 | 0.10 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr8_+_66619277 | 0.10 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr13_+_49684445 | 0.10 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr20_+_56964253 | 0.10 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr16_-_1275257 | 0.10 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr1_+_112016414 | 0.10 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr22_-_18111499 | 0.09 |
ENST00000413576.1
ENST00000399796.2 ENST00000399798.2 ENST00000253413.5 |
ATP6V1E1
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E1 |
| chr12_+_32655048 | 0.09 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr19_+_38880695 | 0.09 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr14_+_56127960 | 0.09 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr13_-_73301819 | 0.09 |
ENST00000377818.3
|
MZT1
|
mitotic spindle organizing protein 1 |
| chr12_+_19358192 | 0.09 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr12_+_51318513 | 0.09 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr1_+_223101757 | 0.09 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr17_-_29641104 | 0.09 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr10_-_99030395 | 0.09 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr17_+_57233087 | 0.09 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr1_+_161736072 | 0.09 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr4_+_175205038 | 0.09 |
ENST00000457424.2
ENST00000514712.1 |
CEP44
|
centrosomal protein 44kDa |
| chr7_+_77469439 | 0.09 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr18_+_32455201 | 0.09 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
| chr9_+_123884038 | 0.09 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr14_+_88490894 | 0.09 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr13_+_53030107 | 0.08 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr12_+_78359999 | 0.08 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr19_+_29456034 | 0.08 |
ENST00000589921.1
|
LINC00906
|
long intergenic non-protein coding RNA 906 |
| chr5_-_115890554 | 0.08 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_+_140800638 | 0.08 |
ENST00000398587.2
ENST00000518882.1 |
PCDHGA11
|
protocadherin gamma subfamily A, 11 |
| chr12_-_118796910 | 0.08 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr15_+_49715293 | 0.08 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr3_+_41241596 | 0.08 |
ENST00000450969.1
|
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr4_-_175443943 | 0.08 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chrX_-_148571884 | 0.08 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr13_+_73302047 | 0.08 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr3_-_197025447 | 0.08 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr2_+_46926048 | 0.08 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr16_-_67965756 | 0.08 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr1_-_54411255 | 0.08 |
ENST00000371377.3
|
HSPB11
|
heat shock protein family B (small), member 11 |
| chr9_-_138391692 | 0.08 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr9_+_20927764 | 0.08 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr10_-_63995871 | 0.08 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr7_-_91794590 | 0.08 |
ENST00000430130.2
ENST00000454089.2 |
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr6_-_43423308 | 0.08 |
ENST00000372485.1
ENST00000372488.3 |
DLK2
|
delta-like 2 homolog (Drosophila) |
| chr21_-_43735446 | 0.08 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr15_+_52121822 | 0.08 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr1_-_86848760 | 0.07 |
ENST00000460698.2
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr6_+_76599809 | 0.07 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr4_-_73935409 | 0.07 |
ENST00000507544.2
ENST00000295890.4 |
COX18
|
COX18 cytochrome C oxidase assembly factor |
| chr2_+_161993412 | 0.07 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr18_+_3449330 | 0.07 |
ENST00000549253.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr4_+_154073469 | 0.07 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr6_+_88299833 | 0.07 |
ENST00000392844.3
ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3
|
origin recognition complex, subunit 3 |
| chr8_+_104831472 | 0.07 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr12_-_11244912 | 0.07 |
ENST00000531678.1
|
TAS2R43
|
taste receptor, type 2, member 43 |
| chr6_+_126661253 | 0.07 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr4_+_156824840 | 0.07 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chrX_-_131547596 | 0.07 |
ENST00000538204.1
ENST00000370849.3 |
MBNL3
|
muscleblind-like splicing regulator 3 |
| chr13_+_78109884 | 0.07 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr21_+_30672433 | 0.07 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr7_-_20256965 | 0.07 |
ENST00000400331.5
ENST00000332878.4 |
MACC1
|
metastasis associated in colon cancer 1 |
| chr9_-_21335240 | 0.07 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr1_-_93257951 | 0.07 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr6_+_143999072 | 0.07 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr2_-_203735586 | 0.07 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr22_+_18111594 | 0.07 |
ENST00000399782.1
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr6_-_134373732 | 0.07 |
ENST00000275230.5
|
SLC2A12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chrX_+_107288280 | 0.07 |
ENST00000458383.1
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr2_+_181988560 | 0.07 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr7_+_150725510 | 0.07 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chrX_-_15683147 | 0.07 |
ENST00000380342.3
|
TMEM27
|
transmembrane protein 27 |
| chr8_-_117886955 | 0.06 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr2_+_233527443 | 0.06 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr16_-_11876408 | 0.06 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr11_+_112047087 | 0.06 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr5_-_79946820 | 0.06 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr2_-_207078154 | 0.06 |
ENST00000447845.1
|
GPR1
|
G protein-coupled receptor 1 |
| chr17_+_40996590 | 0.06 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr1_-_28384598 | 0.06 |
ENST00000373864.1
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr12_+_16500037 | 0.06 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr5_+_56471592 | 0.06 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr3_-_100712292 | 0.06 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr19_-_51531210 | 0.06 |
ENST00000391804.3
|
KLK11
|
kallikrein-related peptidase 11 |
| chr17_+_41005283 | 0.06 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr18_-_61329118 | 0.06 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr2_+_136343904 | 0.06 |
ENST00000436436.1
|
R3HDM1
|
R3H domain containing 1 |
| chr20_-_17539456 | 0.06 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr17_+_28705921 | 0.06 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr10_+_85933494 | 0.06 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr5_+_108083517 | 0.06 |
ENST00000281092.4
ENST00000536402.1 |
FER
|
fer (fps/fes related) tyrosine kinase |
| chr12_+_19358228 | 0.06 |
ENST00000424268.1
ENST00000543806.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr11_-_61687739 | 0.06 |
ENST00000531922.1
ENST00000301773.5 |
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_+_179135246 | 0.06 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr2_-_37544209 | 0.06 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr4_-_152149033 | 0.06 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr16_-_15950868 | 0.06 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr14_+_58894706 | 0.06 |
ENST00000261244.5
|
KIAA0586
|
KIAA0586 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 0.7 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.2 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.3 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.4 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.3 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.4 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0060467 | negative regulation of fertilization(GO:0060467) regulation of binding of sperm to zona pellucida(GO:2000359) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.2 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0007403 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0050904 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.0 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0004423 | iduronate-2-sulfatase activity(GO:0004423) |
| 0.0 | 0.2 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |