Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CDC5L
Z-value: 0.63
Transcription factors associated with CDC5L
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDC5L
|
ENSG00000096401.7 | cell division cycle 5 like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDC5L | hg19_v2_chr6_+_44355257_44355315 | -0.92 | 9.6e-03 | Click! |
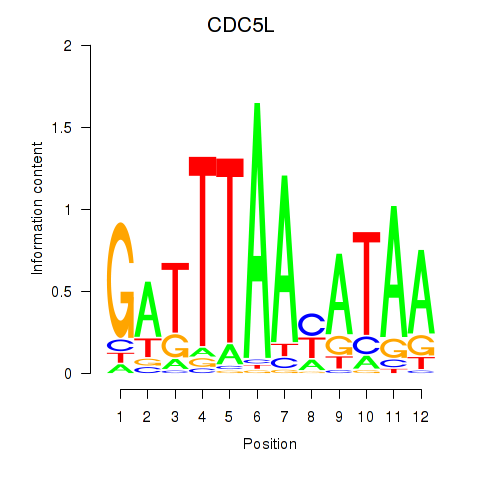
Activity profile of CDC5L motif
Sorted Z-values of CDC5L motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26250835 | 0.82 |
ENST00000446824.2
|
HIST1H3F
|
histone cluster 1, H3f |
| chr14_+_39703112 | 0.73 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr2_+_228678550 | 0.54 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr15_-_42076229 | 0.52 |
ENST00000597767.1
|
AC073657.1
|
Uncharacterized protein |
| chr10_-_69597828 | 0.47 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_-_166702601 | 0.47 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr6_+_27791862 | 0.42 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr4_+_74606223 | 0.42 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr6_+_26156551 | 0.38 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr6_+_32812568 | 0.37 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr10_-_69597915 | 0.36 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr12_-_25150373 | 0.35 |
ENST00000549828.1
|
C12orf77
|
chromosome 12 open reading frame 77 |
| chr11_+_64794991 | 0.35 |
ENST00000352068.5
ENST00000525648.1 |
SNX15
|
sorting nexin 15 |
| chr14_-_38036271 | 0.32 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr14_+_29241910 | 0.30 |
ENST00000399387.4
ENST00000552957.1 ENST00000548213.1 |
C14orf23
|
chromosome 14 open reading frame 23 |
| chr3_+_73110810 | 0.29 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chrY_+_15418467 | 0.29 |
ENST00000595988.1
|
AC010877.1
|
Uncharacterized protein |
| chr10_+_91152303 | 0.28 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr7_+_18536090 | 0.27 |
ENST00000441986.1
|
HDAC9
|
histone deacetylase 9 |
| chr6_-_133055815 | 0.27 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr18_+_3466248 | 0.27 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr1_+_10509971 | 0.26 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr6_-_133055896 | 0.26 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr1_+_152956549 | 0.26 |
ENST00000307122.2
|
SPRR1A
|
small proline-rich protein 1A |
| chr3_+_177545563 | 0.25 |
ENST00000434309.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr19_+_40873617 | 0.25 |
ENST00000599353.1
|
PLD3
|
phospholipase D family, member 3 |
| chr11_+_64794875 | 0.24 |
ENST00000377244.3
ENST00000534637.1 ENST00000524831.1 |
SNX15
|
sorting nexin 15 |
| chr9_-_21368075 | 0.24 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr1_-_153283194 | 0.24 |
ENST00000290722.1
|
PGLYRP3
|
peptidoglycan recognition protein 3 |
| chr6_-_152623231 | 0.23 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_+_229440129 | 0.22 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr4_+_75023816 | 0.21 |
ENST00000395759.2
ENST00000331145.6 ENST00000359107.5 ENST00000325278.6 |
MTHFD2L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr3_-_178865747 | 0.21 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr19_+_782755 | 0.21 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr2_+_232573222 | 0.21 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr2_+_232573208 | 0.20 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr18_+_61554932 | 0.20 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr8_+_86999516 | 0.20 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr1_+_81001398 | 0.20 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr11_-_62609281 | 0.20 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr5_+_43033818 | 0.19 |
ENST00000607830.1
|
CTD-2035E11.4
|
CTD-2035E11.4 |
| chr5_+_140514782 | 0.19 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr3_-_57326704 | 0.19 |
ENST00000487349.1
ENST00000389601.3 |
ASB14
|
ankyrin repeat and SOCS box containing 14 |
| chr19_-_48759119 | 0.19 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr19_-_4540486 | 0.19 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr10_-_28623368 | 0.19 |
ENST00000441595.2
|
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_89591749 | 0.19 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr3_+_151531859 | 0.18 |
ENST00000488869.1
|
AADAC
|
arylacetamide deacetylase |
| chr16_+_33204156 | 0.18 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr10_+_124739964 | 0.18 |
ENST00000406217.2
|
PSTK
|
phosphoseryl-tRNA kinase |
| chr9_-_130890662 | 0.18 |
ENST00000277462.5
ENST00000338961.6 |
PTGES2
|
prostaglandin E synthase 2 |
| chrX_-_15402498 | 0.18 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr1_+_43613612 | 0.18 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr18_+_72168325 | 0.18 |
ENST00000582666.1
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr8_+_99956759 | 0.18 |
ENST00000522510.1
ENST00000457907.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr6_+_41604620 | 0.18 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr18_+_68002675 | 0.18 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr8_+_38065104 | 0.18 |
ENST00000521311.1
|
BAG4
|
BCL2-associated athanogene 4 |
| chr13_+_78315528 | 0.18 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr16_+_4845379 | 0.18 |
ENST00000588606.1
ENST00000586005.1 |
SMIM22
|
small integral membrane protein 22 |
| chr2_+_132479948 | 0.17 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr13_+_78315348 | 0.17 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr19_+_46498704 | 0.17 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr7_-_143956815 | 0.17 |
ENST00000493325.1
|
OR2A7
|
olfactory receptor, family 2, subfamily A, member 7 |
| chr10_+_51549498 | 0.17 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr5_+_139505520 | 0.16 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr4_-_70653673 | 0.16 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr22_-_19466643 | 0.16 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr18_+_616672 | 0.16 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr9_+_105757590 | 0.16 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr12_-_49463753 | 0.16 |
ENST00000301068.6
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr15_+_81299370 | 0.16 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr19_+_51897742 | 0.16 |
ENST00000600765.1
|
CTD-2616J11.14
|
CTD-2616J11.14 |
| chr3_-_48732926 | 0.15 |
ENST00000453202.1
|
IP6K2
|
inositol hexakisphosphate kinase 2 |
| chr19_-_48753028 | 0.15 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr6_-_24489842 | 0.15 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr6_-_131211534 | 0.15 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr6_+_126221034 | 0.15 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr6_-_27840099 | 0.14 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr10_+_69865866 | 0.13 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr4_-_110723134 | 0.13 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr22_+_30821732 | 0.13 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr1_-_154178803 | 0.13 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr5_+_53686658 | 0.13 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr13_+_111855399 | 0.13 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr10_+_46994087 | 0.13 |
ENST00000374317.1
|
GPRIN2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr3_+_8543393 | 0.13 |
ENST00000157600.3
ENST00000415597.1 ENST00000535732.1 |
LMCD1
|
LIM and cysteine-rich domains 1 |
| chr9_-_136214877 | 0.13 |
ENST00000446777.1
ENST00000343730.5 ENST00000344469.5 ENST00000371999.1 ENST00000494177.2 ENST00000457204.2 |
MED22
|
mediator complex subunit 22 |
| chr21_+_33784670 | 0.13 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr2_+_90248739 | 0.13 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr10_-_74114714 | 0.13 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr2_+_166095898 | 0.13 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr15_+_54901540 | 0.12 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr22_-_19466454 | 0.12 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr3_-_119396193 | 0.12 |
ENST00000484810.1
ENST00000497116.1 ENST00000261070.2 |
COX17
|
COX17 cytochrome c oxidase copper chaperone |
| chr6_+_161123270 | 0.12 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chrX_+_10126488 | 0.12 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr21_+_17792672 | 0.12 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr12_-_91546926 | 0.12 |
ENST00000550758.1
|
DCN
|
decorin |
| chr1_-_231560790 | 0.12 |
ENST00000366641.3
|
EGLN1
|
egl-9 family hypoxia-inducible factor 1 |
| chr10_-_69597810 | 0.12 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_-_106098162 | 0.12 |
ENST00000337478.1
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr4_+_147145709 | 0.12 |
ENST00000504313.1
|
RP11-6L6.2
|
Uncharacterized protein |
| chr11_+_2415061 | 0.12 |
ENST00000481687.1
|
CD81
|
CD81 molecule |
| chr7_-_14880892 | 0.12 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr19_+_107471 | 0.12 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr12_-_111358372 | 0.12 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr18_+_616711 | 0.12 |
ENST00000579494.1
|
CLUL1
|
clusterin-like 1 (retinal) |
| chr11_-_83984231 | 0.12 |
ENST00000330014.6
ENST00000537455.1 ENST00000376106.3 ENST00000418306.2 ENST00000531015.1 |
DLG2
|
discs, large homolog 2 (Drosophila) |
| chr2_-_106013207 | 0.11 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr11_+_114168773 | 0.11 |
ENST00000542647.1
ENST00000545255.1 |
NNMT
|
nicotinamide N-methyltransferase |
| chr8_+_99956662 | 0.11 |
ENST00000523368.1
ENST00000297565.4 ENST00000435298.2 |
OSR2
|
odd-skipped related transciption factor 2 |
| chr6_+_15401075 | 0.11 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr17_+_45908974 | 0.11 |
ENST00000269025.4
|
LRRC46
|
leucine rich repeat containing 46 |
| chr8_+_24772455 | 0.11 |
ENST00000433454.2
|
NEFM
|
neurofilament, medium polypeptide |
| chr12_-_10542617 | 0.11 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr2_-_175712479 | 0.11 |
ENST00000443238.1
|
CHN1
|
chimerin 1 |
| chr17_+_73452695 | 0.11 |
ENST00000582186.1
ENST00000582455.1 ENST00000581252.1 ENST00000579208.1 |
KIAA0195
|
KIAA0195 |
| chr20_+_1246908 | 0.11 |
ENST00000381873.3
ENST00000381867.1 |
SNPH
|
syntaphilin |
| chr15_+_67841330 | 0.11 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr21_+_43619796 | 0.11 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr4_+_71384300 | 0.10 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr20_+_44330651 | 0.10 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chr16_+_66600294 | 0.10 |
ENST00000535705.1
ENST00000332695.7 ENST00000336328.6 ENST00000528324.1 ENST00000531885.1 ENST00000529506.1 ENST00000457188.2 ENST00000533666.1 ENST00000533953.1 ENST00000379500.2 ENST00000328020.6 |
CMTM1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr11_-_128737259 | 0.10 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr4_-_110723194 | 0.10 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr16_-_69418553 | 0.10 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr7_-_55620433 | 0.10 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr6_+_30312908 | 0.10 |
ENST00000433076.2
ENST00000442966.2 ENST00000428040.2 ENST00000436442.2 |
RPP21
|
ribonuclease P/MRP 21kDa subunit |
| chr12_+_21525818 | 0.10 |
ENST00000240652.3
ENST00000542023.1 ENST00000537593.1 |
IAPP
|
islet amyloid polypeptide |
| chr4_-_187517928 | 0.10 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr12_-_91573249 | 0.10 |
ENST00000550099.1
ENST00000546391.1 ENST00000551354.1 |
DCN
|
decorin |
| chr9_-_6470375 | 0.10 |
ENST00000355513.4
|
C9orf38
|
chromosome 9 open reading frame 38 |
| chr6_+_27107053 | 0.10 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr3_+_108541545 | 0.10 |
ENST00000295756.6
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1 |
| chr5_+_147582348 | 0.10 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_53948328 | 0.10 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr19_-_1650666 | 0.10 |
ENST00000588136.1
|
TCF3
|
transcription factor 3 |
| chr17_-_7493390 | 0.10 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr16_-_1843694 | 0.10 |
ENST00000569769.1
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr16_-_5116025 | 0.09 |
ENST00000472572.3
ENST00000315997.5 ENST00000422873.1 ENST00000350219.4 |
C16orf89
|
chromosome 16 open reading frame 89 |
| chr3_-_100558953 | 0.09 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr1_+_144339738 | 0.09 |
ENST00000538264.1
|
AL592284.1
|
Protein LOC642441 |
| chr7_+_138943265 | 0.09 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr9_-_28670283 | 0.09 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr1_-_186430222 | 0.09 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr2_+_234160217 | 0.09 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr2_-_113542063 | 0.09 |
ENST00000263339.3
|
IL1A
|
interleukin 1, alpha |
| chr1_+_142618769 | 0.09 |
ENST00000446205.2
ENST00000400755.3 ENST00000412092.2 ENST00000413650.1 |
RP11-417J8.3
|
RP11-417J8.3 |
| chr6_+_131958436 | 0.09 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr11_+_101983176 | 0.09 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr10_+_115511213 | 0.09 |
ENST00000361048.1
|
PLEKHS1
|
pleckstrin homology domain containing, family S member 1 |
| chr19_+_11546093 | 0.09 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr1_-_32169761 | 0.09 |
ENST00000271069.6
|
COL16A1
|
collagen, type XVI, alpha 1 |
| chr19_-_38183201 | 0.09 |
ENST00000590008.1
ENST00000358582.4 |
ZNF781
|
zinc finger protein 781 |
| chr7_+_143806607 | 0.09 |
ENST00000408979.2
|
OR2A2
|
olfactory receptor, family 2, subfamily A, member 2 |
| chr22_-_19466683 | 0.09 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr6_-_31509506 | 0.09 |
ENST00000449757.1
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr11_+_59705928 | 0.09 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr12_-_26278030 | 0.09 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chr19_+_3880581 | 0.09 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr13_+_78315295 | 0.09 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr6_-_41254403 | 0.09 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr16_-_4401258 | 0.09 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr9_-_19786926 | 0.09 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr19_-_38183228 | 0.09 |
ENST00000587199.1
|
ZFP30
|
ZFP30 zinc finger protein |
| chr12_+_1099675 | 0.09 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr7_+_80267973 | 0.08 |
ENST00000394788.3
ENST00000447544.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr12_-_49581152 | 0.08 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_-_168155577 | 0.08 |
ENST00000541354.1
ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr6_+_76599809 | 0.08 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr2_-_74570520 | 0.08 |
ENST00000394019.2
ENST00000346834.4 ENST00000359484.4 ENST00000423644.1 ENST00000377634.4 ENST00000436454.1 |
SLC4A5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr5_+_140579162 | 0.08 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr5_-_68339648 | 0.08 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr2_-_225811747 | 0.08 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr1_-_32169920 | 0.08 |
ENST00000373672.3
ENST00000373668.3 |
COL16A1
|
collagen, type XVI, alpha 1 |
| chr1_-_150738261 | 0.08 |
ENST00000448301.2
ENST00000368985.3 |
CTSS
|
cathepsin S |
| chr12_+_55248289 | 0.08 |
ENST00000308796.6
|
MUCL1
|
mucin-like 1 |
| chr2_+_85921414 | 0.08 |
ENST00000263863.4
ENST00000524600.1 |
GNLY
|
granulysin |
| chr11_-_128712362 | 0.08 |
ENST00000392664.2
|
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr5_+_145316120 | 0.08 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr12_-_58165870 | 0.08 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr13_-_33780133 | 0.08 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr11_+_127140956 | 0.08 |
ENST00000608214.1
|
RP11-480C22.1
|
RP11-480C22.1 |
| chr5_-_95018660 | 0.08 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr7_+_149570049 | 0.08 |
ENST00000421974.2
ENST00000456496.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr22_-_19466732 | 0.08 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr12_+_93096619 | 0.08 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr10_+_81462983 | 0.08 |
ENST00000448135.1
ENST00000429828.1 ENST00000372321.1 |
NUTM2B
|
NUT family member 2B |
| chr2_-_27632390 | 0.08 |
ENST00000350803.4
ENST00000344034.4 |
PPM1G
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr6_-_88001706 | 0.08 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr3_-_152058779 | 0.08 |
ENST00000408960.3
|
TMEM14E
|
transmembrane protein 14E |
| chr10_+_135207598 | 0.08 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr7_-_14026063 | 0.08 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr4_-_186734275 | 0.08 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_189839046 | 0.08 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr5_-_177207634 | 0.08 |
ENST00000513554.1
ENST00000440605.3 |
FAM153A
|
family with sequence similarity 153, member A |
| chr1_+_183605222 | 0.08 |
ENST00000536277.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_+_140180635 | 0.08 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr4_-_23735183 | 0.08 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDC5L
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.2 | GO:0032824 | negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) positive regulation of cytolysis in other organism(GO:0051714) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:1902362 | melanocyte apoptotic process(GO:1902362) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.2 | GO:0010982 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.2 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0033122 | negative regulation of cyclic nucleotide catabolic process(GO:0030806) negative regulation of cAMP catabolic process(GO:0030821) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 0.0 | 0.5 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:2001280 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0070942 | neutrophil mediated cytotoxicity(GO:0070942) neutrophil mediated killing of symbiont cell(GO:0070943) neutrophil mediated killing of bacterium(GO:0070944) neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.1 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.2 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0018315 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:1901896 | PML body organization(GO:0030578) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.5 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.2 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0052836 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |