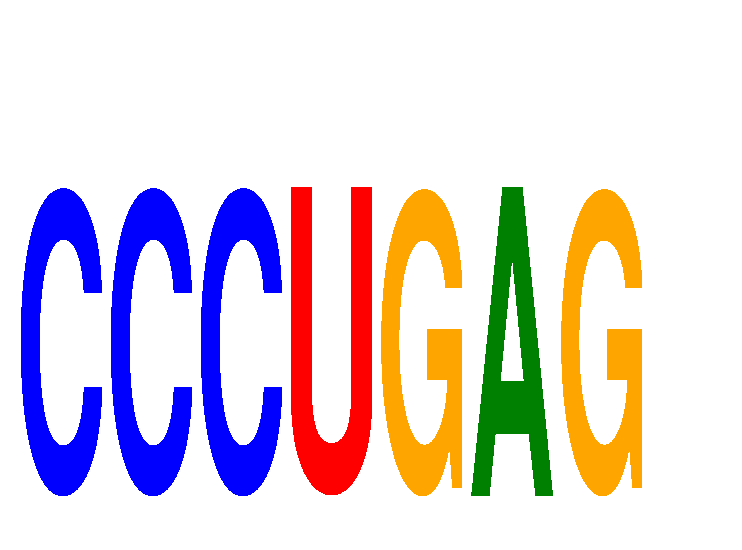Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CCCUGAG
Z-value: 0.63
miRNA associated with seed CCCUGAG
| Name | miRBASE accession |
|---|---|
|
hsa-miR-125a-5p
|
MIMAT0000443 |
|
hsa-miR-125b-5p
|
MIMAT0000423 |
|
hsa-miR-4319
|
MIMAT0016870 |
Activity profile of CCCUGAG motif
Sorted Z-values of CCCUGAG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_138188551 | 0.40 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_+_13906250 | 0.23 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr7_-_131241361 | 0.23 |
ENST00000378555.3
ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL
|
podocalyxin-like |
| chr15_+_90744533 | 0.21 |
ENST00000411539.2
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr8_-_141645645 | 0.21 |
ENST00000519980.1
ENST00000220592.5 |
AGO2
|
argonaute RISC catalytic component 2 |
| chr17_-_61777459 | 0.21 |
ENST00000578993.1
ENST00000583211.1 ENST00000259006.3 |
LIMD2
|
LIM domain containing 2 |
| chr10_+_120967072 | 0.20 |
ENST00000392870.2
|
GRK5
|
G protein-coupled receptor kinase 5 |
| chr11_+_120195992 | 0.20 |
ENST00000314475.2
ENST00000529187.1 |
TMEM136
|
transmembrane protein 136 |
| chr9_-_14314066 | 0.18 |
ENST00000397575.3
|
NFIB
|
nuclear factor I/B |
| chrX_+_28605516 | 0.18 |
ENST00000378993.1
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1 |
| chr19_+_7968728 | 0.17 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr3_-_53381539 | 0.16 |
ENST00000606822.1
ENST00000294241.6 ENST00000607628.1 |
DCP1A
|
decapping mRNA 1A |
| chr1_-_21671968 | 0.15 |
ENST00000415912.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr1_+_165796753 | 0.15 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr3_+_32859510 | 0.15 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr9_+_139981379 | 0.15 |
ENST00000371589.4
|
MAN1B1
|
mannosidase, alpha, class 1B, member 1 |
| chr22_+_21771656 | 0.15 |
ENST00000407464.2
|
HIC2
|
hypermethylated in cancer 2 |
| chr8_-_144699628 | 0.15 |
ENST00000529048.1
ENST00000529064.1 |
TSTA3
|
tissue specific transplantation antigen P35B |
| chr14_+_56585048 | 0.14 |
ENST00000267460.4
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr15_-_100258029 | 0.14 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chr5_-_159739532 | 0.14 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr19_-_18632861 | 0.14 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr19_-_1863567 | 0.13 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr6_-_32811771 | 0.13 |
ENST00000395339.3
ENST00000374882.3 |
PSMB8
|
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr10_+_134000404 | 0.13 |
ENST00000338492.4
ENST00000368629.1 |
DPYSL4
|
dihydropyrimidinase-like 4 |
| chr17_-_57784755 | 0.13 |
ENST00000537860.1
ENST00000393038.2 ENST00000409433.2 |
PTRH2
|
peptidyl-tRNA hydrolase 2 |
| chr11_+_58939965 | 0.13 |
ENST00000227451.3
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr8_+_22857048 | 0.13 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr6_+_106546808 | 0.13 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr11_+_61447845 | 0.13 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr1_-_249120832 | 0.12 |
ENST00000366472.5
|
SH3BP5L
|
SH3-binding domain protein 5-like |
| chr11_-_62494821 | 0.12 |
ENST00000301785.5
|
HNRNPUL2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr3_+_171758344 | 0.12 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr22_-_50699701 | 0.12 |
ENST00000395780.1
|
MAPK12
|
mitogen-activated protein kinase 12 |
| chr2_-_97535708 | 0.12 |
ENST00000305476.5
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr4_-_10118469 | 0.12 |
ENST00000499869.2
|
WDR1
|
WD repeat domain 1 |
| chr17_-_76713100 | 0.12 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr20_+_2673383 | 0.12 |
ENST00000380648.4
ENST00000342725.5 |
EBF4
|
early B-cell factor 4 |
| chr8_-_145550571 | 0.12 |
ENST00000332324.4
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr19_+_45596218 | 0.12 |
ENST00000421905.1
ENST00000221462.4 |
PPP1R37
|
protein phosphatase 1, regulatory subunit 37 |
| chr20_+_62795827 | 0.11 |
ENST00000328439.1
ENST00000536311.1 |
MYT1
|
myelin transcription factor 1 |
| chr17_+_18218587 | 0.11 |
ENST00000406438.3
|
SMCR8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr15_+_31619013 | 0.11 |
ENST00000307145.3
|
KLF13
|
Kruppel-like factor 13 |
| chr9_+_131102925 | 0.11 |
ENST00000372870.1
ENST00000300456.4 |
SLC27A4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr3_+_42695176 | 0.11 |
ENST00000232974.6
ENST00000457842.3 |
ZBTB47
|
zinc finger and BTB domain containing 47 |
| chr19_-_10946949 | 0.11 |
ENST00000214869.2
ENST00000591695.1 |
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr1_+_66999799 | 0.11 |
ENST00000371035.3
ENST00000371036.3 ENST00000371037.4 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr5_-_180288248 | 0.11 |
ENST00000512132.1
ENST00000506439.1 ENST00000502412.1 ENST00000359141.6 |
ZFP62
|
ZFP62 zinc finger protein |
| chr3_+_126707437 | 0.11 |
ENST00000393409.2
ENST00000251772.4 |
PLXNA1
|
plexin A1 |
| chr12_+_122150646 | 0.11 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr1_+_26856236 | 0.11 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr2_+_241375069 | 0.11 |
ENST00000264039.2
|
GPC1
|
glypican 1 |
| chr12_+_69633317 | 0.11 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr2_+_220042933 | 0.11 |
ENST00000430297.2
|
FAM134A
|
family with sequence similarity 134, member A |
| chr3_+_196295482 | 0.10 |
ENST00000440469.1
ENST00000311630.6 |
FBXO45
|
F-box protein 45 |
| chr3_+_47021168 | 0.10 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr16_+_27561449 | 0.10 |
ENST00000261588.4
|
KIAA0556
|
KIAA0556 |
| chr16_+_57126428 | 0.10 |
ENST00000290776.8
|
CPNE2
|
copine II |
| chr7_-_120498357 | 0.10 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chrX_+_110339439 | 0.10 |
ENST00000372010.1
ENST00000519681.1 ENST00000372007.5 |
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr9_-_22009241 | 0.10 |
ENST00000380142.4
|
CDKN2B
|
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr19_+_926000 | 0.10 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_-_65381643 | 0.10 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr3_-_39195037 | 0.10 |
ENST00000273153.5
|
CSRNP1
|
cysteine-serine-rich nuclear protein 1 |
| chr1_+_26737253 | 0.10 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr2_-_213403565 | 0.10 |
ENST00000342788.4
ENST00000436443.1 |
ERBB4
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
| chr3_-_52090461 | 0.09 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr13_-_33859819 | 0.09 |
ENST00000336934.5
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr6_+_7107999 | 0.09 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr8_-_66754172 | 0.09 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr1_+_42846443 | 0.09 |
ENST00000410070.2
ENST00000431473.3 |
RIMKLA
|
ribosomal modification protein rimK-like family member A |
| chr5_-_175964366 | 0.09 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr6_-_31628512 | 0.09 |
ENST00000375911.1
|
C6orf47
|
chromosome 6 open reading frame 47 |
| chr9_+_116638562 | 0.09 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr6_-_16761678 | 0.09 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr14_+_70078303 | 0.09 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr2_+_7057523 | 0.09 |
ENST00000320892.6
|
RNF144A
|
ring finger protein 144A |
| chr20_-_48099182 | 0.09 |
ENST00000371741.4
|
KCNB1
|
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr17_+_2240775 | 0.09 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr17_+_80477571 | 0.09 |
ENST00000335255.5
|
FOXK2
|
forkhead box K2 |
| chr19_-_41196534 | 0.09 |
ENST00000252891.4
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr9_+_77112244 | 0.09 |
ENST00000376896.3
|
RORB
|
RAR-related orphan receptor B |
| chr11_+_61276214 | 0.09 |
ENST00000378075.2
|
LRRC10B
|
leucine rich repeat containing 10B |
| chr4_-_2264015 | 0.09 |
ENST00000337190.2
|
MXD4
|
MAX dimerization protein 4 |
| chr11_+_94277017 | 0.08 |
ENST00000358752.2
|
FUT4
|
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr19_+_19303008 | 0.08 |
ENST00000353145.1
ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK
|
regulatory factor X-associated ankyrin-containing protein |
| chr5_+_139944024 | 0.08 |
ENST00000323146.3
|
SLC35A4
|
solute carrier family 35, member A4 |
| chr15_-_23086394 | 0.08 |
ENST00000337435.4
|
NIPA1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr9_-_19786926 | 0.08 |
ENST00000341998.2
ENST00000286344.3 |
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr6_-_30658745 | 0.08 |
ENST00000376420.5
ENST00000376421.5 |
NRM
|
nurim (nuclear envelope membrane protein) |
| chr3_-_170626418 | 0.08 |
ENST00000474096.1
ENST00000295822.2 |
EIF5A2
|
eukaryotic translation initiation factor 5A2 |
| chr3_-_13461807 | 0.08 |
ENST00000254508.5
|
NUP210
|
nucleoporin 210kDa |
| chr4_-_100575781 | 0.08 |
ENST00000511828.1
|
RP11-766F14.2
|
Protein LOC285556 |
| chr3_-_48672859 | 0.08 |
ENST00000395550.2
ENST00000455886.2 ENST00000431739.1 ENST00000426599.1 ENST00000383733.3 ENST00000420764.2 ENST00000337000.8 |
SLC26A6
|
solute carrier family 26 (anion exchanger), member 6 |
| chr8_-_63998590 | 0.08 |
ENST00000260116.4
|
TTPA
|
tocopherol (alpha) transfer protein |
| chr1_+_29563011 | 0.08 |
ENST00000345512.3
ENST00000373779.3 ENST00000356870.3 ENST00000323874.8 ENST00000428026.2 ENST00000460170.2 |
PTPRU
|
protein tyrosine phosphatase, receptor type, U |
| chr17_+_38375574 | 0.08 |
ENST00000323571.4
ENST00000585043.1 ENST00000394103.3 ENST00000536600.1 |
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr11_-_78052923 | 0.08 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr7_-_143059845 | 0.08 |
ENST00000443739.2
|
FAM131B
|
family with sequence similarity 131, member B |
| chrX_-_103401649 | 0.08 |
ENST00000357421.4
|
SLC25A53
|
solute carrier family 25, member 53 |
| chr20_-_31071239 | 0.08 |
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr18_-_60987220 | 0.08 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr19_-_35454953 | 0.07 |
ENST00000404801.1
|
ZNF792
|
zinc finger protein 792 |
| chr22_+_30279144 | 0.07 |
ENST00000401950.2
ENST00000333027.3 ENST00000445401.1 ENST00000323630.5 ENST00000351488.3 |
MTMR3
|
myotubularin related protein 3 |
| chrX_+_150151752 | 0.07 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr1_-_6321035 | 0.07 |
ENST00000377893.2
|
GPR153
|
G protein-coupled receptor 153 |
| chr17_-_40761375 | 0.07 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr18_+_55102917 | 0.07 |
ENST00000491143.2
|
ONECUT2
|
one cut homeobox 2 |
| chr6_-_11232891 | 0.07 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr15_-_59665062 | 0.07 |
ENST00000288235.4
|
MYO1E
|
myosin IE |
| chr10_+_82213904 | 0.07 |
ENST00000429989.3
|
TSPAN14
|
tetraspanin 14 |
| chr16_+_4897632 | 0.07 |
ENST00000262376.6
|
UBN1
|
ubinuclein 1 |
| chr1_-_45672221 | 0.07 |
ENST00000359600.5
|
ZSWIM5
|
zinc finger, SWIM-type containing 5 |
| chr12_-_120806960 | 0.07 |
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1 |
| chr17_-_42908155 | 0.07 |
ENST00000426548.1
ENST00000590758.1 ENST00000591424.1 |
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr15_-_90358048 | 0.07 |
ENST00000300060.6
ENST00000560137.1 |
ANPEP
|
alanyl (membrane) aminopeptidase |
| chr6_-_31830655 | 0.07 |
ENST00000375631.4
|
NEU1
|
sialidase 1 (lysosomal sialidase) |
| chr18_-_45456930 | 0.07 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr1_+_47901689 | 0.07 |
ENST00000334793.5
|
FOXD2
|
forkhead box D2 |
| chr6_+_17281573 | 0.07 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr2_+_18059906 | 0.07 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr15_+_68871308 | 0.07 |
ENST00000261861.5
|
CORO2B
|
coronin, actin binding protein, 2B |
| chr2_+_124782857 | 0.07 |
ENST00000431078.1
|
CNTNAP5
|
contactin associated protein-like 5 |
| chr2_-_31360887 | 0.07 |
ENST00000420311.2
ENST00000356174.3 ENST00000324589.5 |
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr11_-_64901978 | 0.07 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr2_+_28615669 | 0.07 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr3_+_9773409 | 0.07 |
ENST00000433861.2
ENST00000424362.1 ENST00000383829.2 ENST00000302054.3 ENST00000420291.1 |
BRPF1
|
bromodomain and PHD finger containing, 1 |
| chr19_+_55851221 | 0.07 |
ENST00000255613.3
ENST00000539076.1 |
SUV420H2
AC020922.1
|
suppressor of variegation 4-20 homolog 2 (Drosophila) Uncharacterized protein |
| chr2_+_166326157 | 0.07 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr10_-_94003003 | 0.07 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_-_163695128 | 0.07 |
ENST00000332142.5
|
KCNH7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr11_+_33278811 | 0.07 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr7_+_49813255 | 0.06 |
ENST00000340652.4
|
VWC2
|
von Willebrand factor C domain containing 2 |
| chr19_+_10654561 | 0.06 |
ENST00000309469.4
|
ATG4D
|
autophagy related 4D, cysteine peptidase |
| chr17_-_27224621 | 0.06 |
ENST00000394906.2
ENST00000585169.1 ENST00000394908.4 |
FLOT2
|
flotillin 2 |
| chr14_+_94492674 | 0.06 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr12_-_125348448 | 0.06 |
ENST00000339570.5
|
SCARB1
|
scavenger receptor class B, member 1 |
| chr10_+_97803151 | 0.06 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr12_-_50222187 | 0.06 |
ENST00000335999.6
|
NCKAP5L
|
NCK-associated protein 5-like |
| chr6_-_40555176 | 0.06 |
ENST00000338305.6
|
LRFN2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr2_+_232826211 | 0.06 |
ENST00000325385.7
ENST00000360410.4 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr1_+_197881592 | 0.06 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr5_+_57878859 | 0.06 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr18_-_44702668 | 0.06 |
ENST00000256433.3
|
IER3IP1
|
immediate early response 3 interacting protein 1 |
| chr6_-_100912785 | 0.06 |
ENST00000369208.3
|
SIM1
|
single-minded family bHLH transcription factor 1 |
| chr9_-_130341268 | 0.06 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr17_+_61699766 | 0.06 |
ENST00000579585.1
ENST00000584573.1 ENST00000361733.3 ENST00000361357.3 |
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3 |
| chrX_-_48755030 | 0.06 |
ENST00000490755.2
ENST00000465150.2 ENST00000495490.2 |
TIMM17B
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr11_-_118023490 | 0.06 |
ENST00000324727.4
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr12_+_5019061 | 0.06 |
ENST00000382545.3
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) |
| chr17_-_78009647 | 0.06 |
ENST00000310924.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr10_+_104678032 | 0.06 |
ENST00000369878.4
ENST00000369875.3 |
CNNM2
|
cyclin M2 |
| chr19_+_4969116 | 0.06 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr20_-_33460621 | 0.06 |
ENST00000427420.1
ENST00000336431.5 |
GGT7
|
gamma-glutamyltransferase 7 |
| chr20_+_39765581 | 0.06 |
ENST00000244007.3
|
PLCG1
|
phospholipase C, gamma 1 |
| chr11_-_22851367 | 0.06 |
ENST00000354193.4
|
SVIP
|
small VCP/p97-interacting protein |
| chr22_+_41697520 | 0.06 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr8_-_23712312 | 0.06 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr1_-_179198702 | 0.06 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr19_-_15560730 | 0.06 |
ENST00000389282.4
ENST00000263381.7 |
WIZ
|
widely interspaced zinc finger motifs |
| chr2_+_12857015 | 0.06 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr15_-_93199069 | 0.06 |
ENST00000327355.5
|
FAM174B
|
family with sequence similarity 174, member B |
| chr2_+_47168313 | 0.05 |
ENST00000319190.5
ENST00000394850.2 ENST00000536057.1 |
TTC7A
|
tetratricopeptide repeat domain 7A |
| chr17_+_27920486 | 0.05 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr12_+_49932886 | 0.05 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr3_+_49591881 | 0.05 |
ENST00000296452.4
|
BSN
|
bassoon presynaptic cytomatrix protein |
| chr9_+_35538616 | 0.05 |
ENST00000455600.1
|
RUSC2
|
RUN and SH3 domain containing 2 |
| chr1_-_43833628 | 0.05 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr8_-_116681221 | 0.05 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr2_+_85981008 | 0.05 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr12_-_117319236 | 0.05 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr3_-_49823941 | 0.05 |
ENST00000321599.4
ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1
|
inositol hexakisphosphate kinase 1 |
| chr17_+_21279509 | 0.05 |
ENST00000583088.1
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr9_-_139137648 | 0.05 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr16_+_71929397 | 0.05 |
ENST00000537613.1
ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1
|
increased sodium tolerance 1 homolog (yeast) |
| chrX_+_47077632 | 0.05 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr19_-_49944806 | 0.05 |
ENST00000221485.3
|
SLC17A7
|
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr7_-_98741642 | 0.05 |
ENST00000361368.2
|
SMURF1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr9_+_12775011 | 0.05 |
ENST00000319264.3
|
LURAP1L
|
leucine rich adaptor protein 1-like |
| chr20_-_4982132 | 0.05 |
ENST00000338244.1
ENST00000424750.2 |
SLC23A2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr17_-_48227877 | 0.05 |
ENST00000316878.6
|
PPP1R9B
|
protein phosphatase 1, regulatory subunit 9B |
| chr2_-_201936302 | 0.05 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr16_+_1662326 | 0.05 |
ENST00000397412.3
|
CRAMP1L
|
Crm, cramped-like (Drosophila) |
| chr17_+_61086917 | 0.05 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr8_-_67579418 | 0.05 |
ENST00000310421.4
|
VCPIP1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr1_-_217262969 | 0.05 |
ENST00000361525.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr14_-_23504087 | 0.05 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr5_-_100238956 | 0.05 |
ENST00000231461.5
|
ST8SIA4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr11_+_73882144 | 0.05 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr11_+_66025167 | 0.05 |
ENST00000394067.2
ENST00000316924.5 ENST00000421552.1 ENST00000394078.1 |
KLC2
|
kinesin light chain 2 |
| chr3_+_5229356 | 0.05 |
ENST00000256497.4
|
EDEM1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr8_-_53322303 | 0.05 |
ENST00000276480.7
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein) |
| chr14_-_67982146 | 0.05 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr17_+_72983674 | 0.05 |
ENST00000337231.5
|
CDR2L
|
cerebellar degeneration-related protein 2-like |
| chr19_+_18794470 | 0.05 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr1_+_40974431 | 0.04 |
ENST00000296380.4
ENST00000432259.1 ENST00000418186.1 |
EXO5
|
exonuclease 5 |
| chr17_-_27507395 | 0.04 |
ENST00000354329.4
ENST00000527372.1 |
MYO18A
|
myosin XVIIIA |
| chr9_-_95432536 | 0.04 |
ENST00000287996.3
|
IPPK
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr7_-_75988321 | 0.04 |
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chr2_+_177053307 | 0.04 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CCCUGAG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of osteoclast proliferation(GO:0090291) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0099542 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.2 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.0 | 0.1 | GO:0061026 | central nervous system morphogenesis(GO:0021551) cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0016334 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.2 | GO:0035087 | siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.2 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1904562 | phosphatidylinositol 5-phosphate metabolic process(GO:1904562) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.0 | GO:0070904 | L-ascorbic acid transport(GO:0015882) molecular hydrogen transport(GO:0015993) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0090210 | vitamin E metabolic process(GO:0042360) regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.0 | GO:1901228 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.0 | GO:0045082 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-10 biosynthetic process(GO:0045082) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.0 | GO:0061386 | noradrenergic neuron differentiation(GO:0003357) closure of optic fissure(GO:0061386) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0042670 | amacrine cell differentiation(GO:0035881) retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0043049 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) optic placode formation involved in camera-type eye formation(GO:0046619) branching involved in pancreas morphogenesis(GO:0061114) negative regulation of bile acid biosynthetic process(GO:0070858) acinar cell differentiation(GO:0090425) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.0 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.1 | GO:0050577 | GDP-4-dehydro-D-rhamnose reductase activity(GO:0042356) GDP-L-fucose synthase activity(GO:0050577) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.1 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.0 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0052839 | inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |