Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
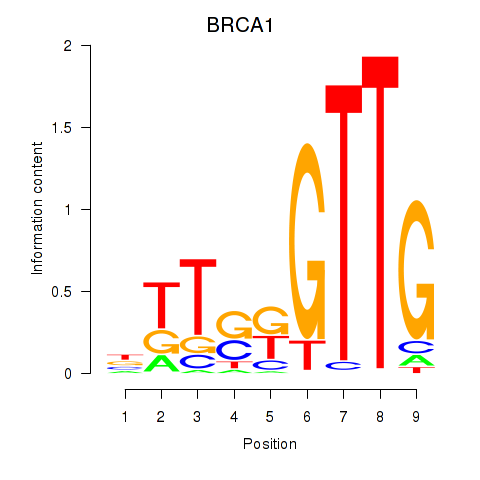
Results for BRCA1
Z-value: 0.40
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277370_41277419 | 0.67 | 1.5e-01 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_97773202 | 0.21 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr8_+_97773457 | 0.15 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr2_+_196522032 | 0.14 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr12_-_96336369 | 0.14 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chrX_-_46187069 | 0.10 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr4_+_619386 | 0.10 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr4_+_619347 | 0.10 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr2_+_223725723 | 0.10 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr20_+_9049682 | 0.09 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr20_+_52105495 | 0.09 |
ENST00000439873.2
|
AL354993.1
|
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chrX_+_49644470 | 0.09 |
ENST00000508866.2
|
USP27X
|
ubiquitin specific peptidase 27, X-linked |
| chr3_-_176914191 | 0.09 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_-_173020056 | 0.09 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr3_-_27410847 | 0.09 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr3_-_176914238 | 0.08 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr2_-_25194476 | 0.08 |
ENST00000534855.1
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr17_-_42019836 | 0.08 |
ENST00000225992.3
|
PPY
|
pancreatic polypeptide |
| chr13_-_50018140 | 0.07 |
ENST00000410043.1
ENST00000347776.5 |
CAB39L
|
calcium binding protein 39-like |
| chr5_+_140079919 | 0.07 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr10_-_95209 | 0.07 |
ENST00000332708.5
ENST00000309812.4 |
TUBB8
|
tubulin, beta 8 class VIII |
| chr2_-_134326009 | 0.07 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr18_-_49557 | 0.06 |
ENST00000308911.6
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr12_+_510795 | 0.06 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr12_-_49418407 | 0.06 |
ENST00000526209.1
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr4_-_140005443 | 0.06 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr2_+_108994466 | 0.06 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr13_-_50018241 | 0.06 |
ENST00000409308.1
|
CAB39L
|
calcium binding protein 39-like |
| chr17_-_7082861 | 0.05 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr5_-_124082279 | 0.05 |
ENST00000513986.1
|
ZNF608
|
zinc finger protein 608 |
| chr9_+_35792151 | 0.05 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr9_-_124976154 | 0.05 |
ENST00000482062.1
|
LHX6
|
LIM homeobox 6 |
| chr19_+_46000479 | 0.05 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr4_-_48782259 | 0.05 |
ENST00000507711.1
ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL
|
FRY-like |
| chr1_+_70687137 | 0.05 |
ENST00000436161.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr4_+_185570767 | 0.05 |
ENST00000314970.6
ENST00000515774.1 ENST00000503752.1 |
PRIMPOL
|
primase and polymerase (DNA-directed) |
| chr12_+_510742 | 0.05 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr10_-_95504 | 0.05 |
ENST00000447903.2
|
TUBB8
|
tubulin, beta 8 class VIII |
| chr13_-_31038370 | 0.05 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr5_+_145583107 | 0.04 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr9_-_86593238 | 0.04 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr1_-_160232291 | 0.04 |
ENST00000368074.1
ENST00000447377.1 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr1_+_159141397 | 0.04 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr5_-_68665084 | 0.04 |
ENST00000509462.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr17_-_8702667 | 0.04 |
ENST00000329805.4
|
MFSD6L
|
major facilitator superfamily domain containing 6-like |
| chr5_-_68665296 | 0.04 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr5_+_145583156 | 0.04 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr6_+_16129308 | 0.04 |
ENST00000356840.3
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr8_-_81083341 | 0.04 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr15_-_72612470 | 0.04 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr5_-_68664989 | 0.04 |
ENST00000508954.1
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr2_+_223725652 | 0.04 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr5_-_68665469 | 0.04 |
ENST00000217893.5
|
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr13_+_27825446 | 0.04 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21 |
| chr5_-_142023766 | 0.03 |
ENST00000394496.2
|
FGF1
|
fibroblast growth factor 1 (acidic) |
| chrX_-_70288234 | 0.03 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr11_-_13484713 | 0.03 |
ENST00000526841.1
ENST00000529708.1 ENST00000278174.5 ENST00000528120.1 |
BTBD10
|
BTB (POZ) domain containing 10 |
| chr14_-_75179774 | 0.03 |
ENST00000555249.1
ENST00000556202.1 ENST00000356357.4 ENST00000338772.5 |
AREL1
AC007956.1
|
apoptosis resistant E3 ubiquitin protein ligase 1 Full-length cDNA 5-PRIME end of clone CS0CAP004YO05 of Thymus of Homo sapiens (human); Uncharacterized protein |
| chr10_+_28822636 | 0.03 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr2_+_98703595 | 0.03 |
ENST00000435344.1
|
VWA3B
|
von Willebrand factor A domain containing 3B |
| chr6_+_42981922 | 0.03 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr7_+_99717230 | 0.03 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr12_-_67072714 | 0.03 |
ENST00000545666.1
ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr5_+_139027877 | 0.03 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr10_-_61900762 | 0.03 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr2_-_152118352 | 0.02 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr12_-_122879969 | 0.02 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr1_-_160232197 | 0.02 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr12_-_95510743 | 0.02 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr6_-_42981651 | 0.02 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr11_+_117947724 | 0.02 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr1_-_160232312 | 0.02 |
ENST00000440682.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr13_+_27825706 | 0.02 |
ENST00000272274.4
ENST00000319826.4 ENST00000326092.4 |
RPL21
|
ribosomal protein L21 |
| chr3_+_185303962 | 0.02 |
ENST00000296257.5
|
SENP2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr14_+_21510385 | 0.02 |
ENST00000298690.4
|
RNASE7
|
ribonuclease, RNase A family, 7 |
| chr1_-_170043709 | 0.02 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr14_+_103058948 | 0.02 |
ENST00000262241.6
|
RCOR1
|
REST corepressor 1 |
| chrX_+_10124977 | 0.02 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr7_+_73868439 | 0.02 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr10_+_92631709 | 0.02 |
ENST00000413330.1
ENST00000277882.3 |
RPP30
|
ribonuclease P/MRP 30kDa subunit |
| chr19_-_37663332 | 0.02 |
ENST00000392157.2
|
ZNF585A
|
zinc finger protein 585A |
| chr11_+_47291645 | 0.02 |
ENST00000395336.3
ENST00000402192.2 |
MADD
|
MAP-kinase activating death domain |
| chr9_-_43133544 | 0.02 |
ENST00000254835.2
|
ANKRD20A3
|
ankyrin repeat domain 20 family, member A3 |
| chr1_+_161736072 | 0.02 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr3_+_187461442 | 0.02 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr11_+_47291193 | 0.02 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr1_+_10271674 | 0.02 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chr18_-_23671139 | 0.01 |
ENST00000579061.1
ENST00000542420.2 |
SS18
|
synovial sarcoma translocation, chromosome 18 |
| chr18_+_19749386 | 0.01 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr1_+_70687079 | 0.01 |
ENST00000395136.2
|
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr11_-_110583912 | 0.01 |
ENST00000533353.1
ENST00000527598.1 |
ARHGAP20
|
Rho GTPase activating protein 20 |
| chr15_-_58358607 | 0.01 |
ENST00000249750.4
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr10_+_102295616 | 0.01 |
ENST00000299163.6
|
HIF1AN
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr1_-_115301235 | 0.01 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chrX_-_10851762 | 0.01 |
ENST00000380785.1
ENST00000380787.1 |
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr8_-_116681221 | 0.01 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr10_+_93683519 | 0.01 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr19_-_6057282 | 0.01 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr12_-_12491608 | 0.01 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr17_-_41739283 | 0.01 |
ENST00000393661.2
ENST00000318579.4 |
MEOX1
|
mesenchyme homeobox 1 |
| chr17_+_36584662 | 0.01 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chrX_-_77150985 | 0.01 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr6_-_170862322 | 0.01 |
ENST00000262193.6
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1 |
| chr7_-_6010263 | 0.00 |
ENST00000455618.2
ENST00000405415.1 ENST00000404406.1 ENST00000542644.1 |
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr2_-_158182105 | 0.00 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr13_-_99910673 | 0.00 |
ENST00000397473.2
ENST00000397470.2 |
GPR18
|
G protein-coupled receptor 18 |
| chr13_+_103046954 | 0.00 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr14_-_94942548 | 0.00 |
ENST00000298845.7
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| chr7_-_78400364 | 0.00 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr15_-_88799948 | 0.00 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr14_-_94942527 | 0.00 |
ENST00000448305.2
|
SERPINA9
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:2000329 | negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.0 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) T-helper 1 cell activation(GO:0035711) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |