Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for BBX
Z-value: 0.53
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.14 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg19_v2_chr3_+_107241783_107241850 | -0.73 | 1.0e-01 | Click! |
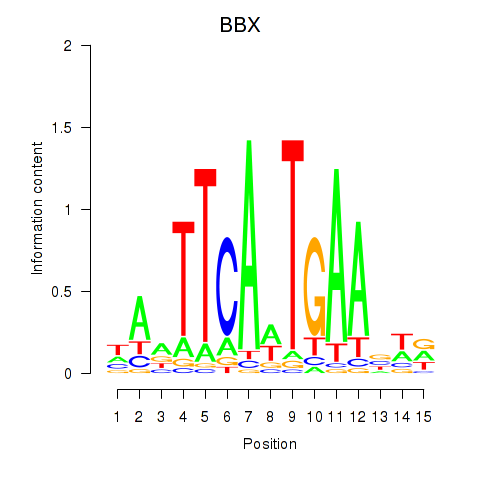
Activity profile of BBX motif
Sorted Z-values of BBX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_119669160 | 0.56 |
ENST00000514240.1
|
CTC-552D5.1
|
CTC-552D5.1 |
| chr11_+_18154059 | 0.56 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr11_-_105010320 | 0.55 |
ENST00000532895.1
ENST00000530950.1 |
CARD18
|
caspase recruitment domain family, member 18 |
| chr6_-_107235331 | 0.37 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr17_+_45331184 | 0.36 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr3_+_177534653 | 0.36 |
ENST00000436078.1
|
RP11-91K9.1
|
RP11-91K9.1 |
| chr5_+_140514782 | 0.31 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr19_+_49109990 | 0.25 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr13_-_81801115 | 0.24 |
ENST00000567258.1
|
LINC00564
|
long intergenic non-protein coding RNA 564 |
| chr14_-_23292596 | 0.24 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr10_-_4285835 | 0.22 |
ENST00000454470.1
|
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr1_+_229440129 | 0.21 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chrX_-_153141783 | 0.20 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr14_+_94492674 | 0.19 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr10_-_4285923 | 0.17 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr2_+_13677795 | 0.15 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr12_-_66035922 | 0.15 |
ENST00000546198.1
ENST00000535315.1 ENST00000537298.1 |
RP11-230G5.2
|
RP11-230G5.2 |
| chr14_+_75230011 | 0.14 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr1_+_28527070 | 0.14 |
ENST00000596102.1
|
AL353354.2
|
AL353354.2 |
| chr14_-_81425828 | 0.14 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr16_-_31564881 | 0.13 |
ENST00000567765.1
|
CTD-2014E2.6
|
CTD-2014E2.6 |
| chr7_-_38969150 | 0.12 |
ENST00000418457.2
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr6_-_133084580 | 0.12 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr3_+_127770455 | 0.12 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr17_+_4853442 | 0.12 |
ENST00000522301.1
|
ENO3
|
enolase 3 (beta, muscle) |
| chr18_+_63273310 | 0.11 |
ENST00000579862.1
|
RP11-775G23.1
|
RP11-775G23.1 |
| chr10_-_94003003 | 0.11 |
ENST00000412050.4
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr16_+_56672571 | 0.11 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr6_+_106534192 | 0.10 |
ENST00000369091.2
ENST00000369096.4 |
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr19_+_31658405 | 0.09 |
ENST00000589511.1
|
CTC-439O9.3
|
CTC-439O9.3 |
| chr2_+_228735763 | 0.09 |
ENST00000373666.2
|
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr2_-_120124258 | 0.09 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chrX_+_54834004 | 0.09 |
ENST00000375068.1
|
MAGED2
|
melanoma antigen family D, 2 |
| chr12_-_66036137 | 0.09 |
ENST00000539653.1
ENST00000544557.1 |
RP11-230G5.2
|
RP11-230G5.2 |
| chr7_+_128349106 | 0.09 |
ENST00000485070.1
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr6_-_107235378 | 0.09 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr4_-_186682716 | 0.09 |
ENST00000445343.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr6_-_88001706 | 0.08 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr1_-_153123345 | 0.08 |
ENST00000368748.4
|
SPRR2G
|
small proline-rich protein 2G |
| chr5_+_40841410 | 0.08 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr2_+_232135245 | 0.08 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr5_+_140729649 | 0.07 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr11_+_61717279 | 0.07 |
ENST00000378043.4
|
BEST1
|
bestrophin 1 |
| chr5_+_140480083 | 0.07 |
ENST00000231130.2
|
PCDHB3
|
protocadherin beta 3 |
| chr7_+_117120017 | 0.07 |
ENST00000003084.6
ENST00000454343.1 |
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr11_+_61717336 | 0.07 |
ENST00000378042.3
|
BEST1
|
bestrophin 1 |
| chr2_+_110656005 | 0.07 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr12_-_49581152 | 0.07 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chrX_+_54834159 | 0.07 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr18_+_29769978 | 0.07 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr14_+_38264418 | 0.06 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chrX_-_19817869 | 0.06 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chrX_-_52827141 | 0.06 |
ENST00000375511.3
|
SPANXN5
|
SPANX family, member N5 |
| chr2_+_113816215 | 0.06 |
ENST00000346807.3
|
IL36RN
|
interleukin 36 receptor antagonist |
| chr1_+_55464600 | 0.06 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr4_-_186696425 | 0.06 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr10_+_24497704 | 0.06 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr7_-_107968999 | 0.06 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr5_+_140810132 | 0.05 |
ENST00000252085.3
|
PCDHGA12
|
protocadherin gamma subfamily A, 12 |
| chr6_+_31895287 | 0.05 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr1_-_161337662 | 0.05 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr15_+_54901540 | 0.05 |
ENST00000539562.2
|
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr13_+_50570019 | 0.05 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr12_-_10978957 | 0.05 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr16_-_58768177 | 0.05 |
ENST00000434819.2
ENST00000245206.5 |
GOT2
|
glutamic-oxaloacetic transaminase 2, mitochondrial |
| chr15_+_54305101 | 0.05 |
ENST00000260323.11
ENST00000545554.1 ENST00000537900.1 |
UNC13C
|
unc-13 homolog C (C. elegans) |
| chr20_-_7921090 | 0.05 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_-_130080977 | 0.05 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr12_-_4754318 | 0.05 |
ENST00000536414.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr12_-_39734783 | 0.05 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr7_+_117120106 | 0.04 |
ENST00000426809.1
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr4_-_186696636 | 0.04 |
ENST00000444771.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_206785789 | 0.04 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr7_+_95171457 | 0.04 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr4_-_186734275 | 0.04 |
ENST00000456060.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_69313145 | 0.04 |
ENST00000305363.4
|
TMPRSS11E
|
transmembrane protease, serine 11E |
| chr6_+_29424958 | 0.04 |
ENST00000377136.1
ENST00000377133.1 |
OR2H1
|
olfactory receptor, family 2, subfamily H, member 1 |
| chr6_-_130543958 | 0.04 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr6_+_31895254 | 0.04 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr6_+_53948328 | 0.04 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr16_-_28374829 | 0.04 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr4_-_104021009 | 0.04 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr4_-_186733363 | 0.04 |
ENST00000393523.2
ENST00000393528.3 ENST00000449407.2 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr9_-_95244781 | 0.03 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr10_+_90562487 | 0.03 |
ENST00000404743.4
|
LIPM
|
lipase, family member M |
| chr17_+_40950900 | 0.03 |
ENST00000588527.1
|
CNTD1
|
cyclin N-terminal domain containing 1 |
| chr4_-_987217 | 0.03 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr11_+_35222629 | 0.03 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr2_-_158182410 | 0.03 |
ENST00000419116.2
ENST00000410096.1 |
ERMN
|
ermin, ERM-like protein |
| chr6_-_49712123 | 0.03 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_+_108910870 | 0.03 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr3_-_47950745 | 0.03 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr20_+_23420885 | 0.03 |
ENST00000246020.2
|
CSTL1
|
cystatin-like 1 |
| chrX_+_144328348 | 0.03 |
ENST00000370493.3
|
SPANXN1
|
SPANX family, member N1 |
| chr2_-_231084617 | 0.03 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr6_+_72922505 | 0.03 |
ENST00000401910.3
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr1_-_59043166 | 0.03 |
ENST00000371225.2
|
TACSTD2
|
tumor-associated calcium signal transducer 2 |
| chr1_-_216978709 | 0.03 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr14_+_23235886 | 0.02 |
ENST00000604262.1
ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L
|
oxidase (cytochrome c) assembly 1-like |
| chr4_-_186697044 | 0.02 |
ENST00000437304.2
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_125758813 | 0.02 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_+_22689648 | 0.02 |
ENST00000278187.3
|
GAS2
|
growth arrest-specific 2 |
| chr6_+_72922590 | 0.02 |
ENST00000523963.1
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
| chr20_-_30310656 | 0.02 |
ENST00000376055.4
|
BCL2L1
|
BCL2-like 1 |
| chr9_-_107361788 | 0.02 |
ENST00000374779.2
|
OR13C5
|
olfactory receptor, family 13, subfamily C, member 5 |
| chr22_+_44319619 | 0.02 |
ENST00000216180.3
|
PNPLA3
|
patatin-like phospholipase domain containing 3 |
| chr3_+_111260856 | 0.02 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr5_+_150040403 | 0.02 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr7_-_121944491 | 0.02 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr12_-_100378006 | 0.02 |
ENST00000547776.2
ENST00000329257.7 ENST00000547010.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_54173227 | 0.02 |
ENST00000259782.4
ENST00000370864.3 |
TINAG
|
tubulointerstitial nephritis antigen |
| chr4_-_186696515 | 0.02 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr20_-_30310693 | 0.02 |
ENST00000307677.4
ENST00000420653.1 |
BCL2L1
|
BCL2-like 1 |
| chr3_+_122399444 | 0.01 |
ENST00000474629.2
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr3_+_140981456 | 0.01 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr4_-_987164 | 0.01 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr20_-_30310336 | 0.01 |
ENST00000434194.1
ENST00000376062.2 |
BCL2L1
|
BCL2-like 1 |
| chr18_-_56985776 | 0.01 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr3_+_111260954 | 0.01 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr19_+_6887571 | 0.01 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chrX_-_119077695 | 0.01 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr4_-_186696561 | 0.01 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_+_101437487 | 0.01 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr5_+_74011328 | 0.01 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr20_-_56286479 | 0.00 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr16_-_8622226 | 0.00 |
ENST00000568335.1
|
TMEM114
|
transmembrane protein 114 |
| chr4_-_103998060 | 0.00 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr2_-_158182322 | 0.00 |
ENST00000420719.2
ENST00000409216.1 |
ERMN
|
ermin, ERM-like protein |
| chr9_+_35161998 | 0.00 |
ENST00000396787.1
ENST00000378495.3 ENST00000378496.4 |
UNC13B
|
unc-13 homolog B (C. elegans) |
| chr13_+_76413852 | 0.00 |
ENST00000533809.2
|
LMO7
|
LIM domain 7 |
| chr17_-_17184605 | 0.00 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chr4_-_104020968 | 0.00 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr8_-_108510224 | 0.00 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.6 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.1 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |