Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
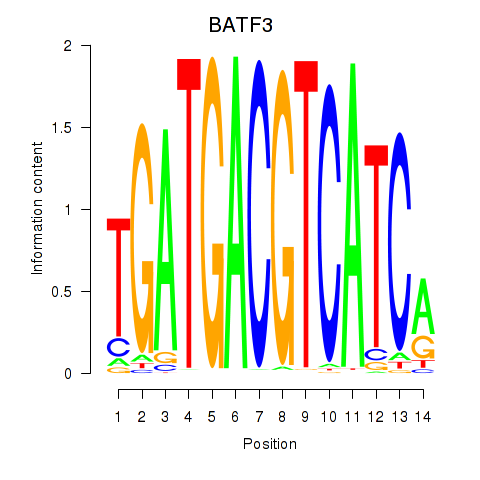
Results for BATF3
Z-value: 0.31
Transcription factors associated with BATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BATF3
|
ENSG00000123685.4 | basic leucine zipper ATF-like transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BATF3 | hg19_v2_chr1_-_212873267_212873332 | 0.27 | 6.0e-01 | Click! |
Activity profile of BATF3 motif
Sorted Z-values of BATF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_49622348 | 0.29 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr11_+_102217936 | 0.16 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr7_-_93633684 | 0.13 |
ENST00000222547.3
ENST00000425626.1 |
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr15_+_96904487 | 0.12 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr22_+_38597889 | 0.11 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr22_+_31518938 | 0.09 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr14_-_24732738 | 0.09 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr17_-_48277552 | 0.09 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr14_-_24732403 | 0.08 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr14_-_24732368 | 0.08 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr14_-_35182994 | 0.08 |
ENST00000341223.3
|
CFL2
|
cofilin 2 (muscle) |
| chr3_-_134092561 | 0.08 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr3_+_184056614 | 0.08 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr19_+_47616682 | 0.07 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr9_-_100684769 | 0.07 |
ENST00000455506.1
ENST00000375117.4 |
C9orf156
|
chromosome 9 open reading frame 156 |
| chrX_-_135056106 | 0.07 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr12_+_10365082 | 0.07 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr6_+_31939608 | 0.07 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr20_+_42219559 | 0.07 |
ENST00000373030.3
ENST00000373039.4 |
IFT52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr17_-_49124230 | 0.06 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr2_-_98280383 | 0.06 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chrX_-_135056216 | 0.06 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr5_-_172198190 | 0.06 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr1_-_70671216 | 0.05 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr2_+_220071490 | 0.05 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr19_+_8455200 | 0.05 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr3_-_119182506 | 0.05 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr2_+_70142189 | 0.05 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr11_-_28129656 | 0.04 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr2_-_43823119 | 0.04 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr1_+_228353495 | 0.04 |
ENST00000366711.3
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr22_+_37959647 | 0.04 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr16_+_3068393 | 0.04 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr3_-_52312337 | 0.04 |
ENST00000469000.1
|
WDR82
|
WD repeat domain 82 |
| chr12_-_92539614 | 0.04 |
ENST00000256015.3
|
BTG1
|
B-cell translocation gene 1, anti-proliferative |
| chr7_-_93633658 | 0.04 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr16_+_56225248 | 0.03 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr2_-_65357225 | 0.03 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr17_+_7835419 | 0.03 |
ENST00000576538.1
ENST00000380262.3 ENST00000563694.1 ENST00000380255.3 ENST00000570782.1 |
CNTROB
|
centrobin, centrosomal BRCA2 interacting protein |
| chr19_-_47616992 | 0.03 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr2_+_70142232 | 0.03 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr11_+_12695944 | 0.03 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr3_-_101405539 | 0.03 |
ENST00000469605.1
ENST00000495401.1 ENST00000394077.3 |
RPL24
|
ribosomal protein L24 |
| chr7_+_100464760 | 0.03 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr13_-_36871886 | 0.03 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr1_+_156030937 | 0.02 |
ENST00000361084.5
|
RAB25
|
RAB25, member RAS oncogene family |
| chr8_+_37594185 | 0.02 |
ENST00000518586.1
ENST00000335171.6 ENST00000521644.1 |
ERLIN2
|
ER lipid raft associated 2 |
| chr2_+_74757050 | 0.02 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr8_+_37594130 | 0.02 |
ENST00000518526.1
ENST00000523887.1 ENST00000276461.5 |
ERLIN2
|
ER lipid raft associated 2 |
| chr5_-_175964366 | 0.01 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr2_-_43823093 | 0.01 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr12_+_10365404 | 0.01 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr15_+_42841008 | 0.01 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr2_-_25194963 | 0.01 |
ENST00000264711.2
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr11_+_12696102 | 0.01 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr11_+_28129795 | 0.01 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr15_-_34394119 | 0.01 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr5_-_175965008 | 0.01 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr1_+_153750622 | 0.01 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr3_-_52312636 | 0.01 |
ENST00000296490.3
|
WDR82
|
WD repeat domain 82 |
| chr6_-_31940065 | 0.00 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr4_-_104119488 | 0.00 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr4_-_104119528 | 0.00 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr3_+_38206975 | 0.00 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr17_+_29421900 | 0.00 |
ENST00000358273.4
ENST00000356175.3 |
NF1
|
neurofibromin 1 |
| chr11_+_66059339 | 0.00 |
ENST00000327259.4
|
TMEM151A
|
transmembrane protein 151A |
| chr16_+_68056844 | 0.00 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BATF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.3 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |