Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for BARHL2
Z-value: 0.31
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.7 | BarH like homeobox 2 |
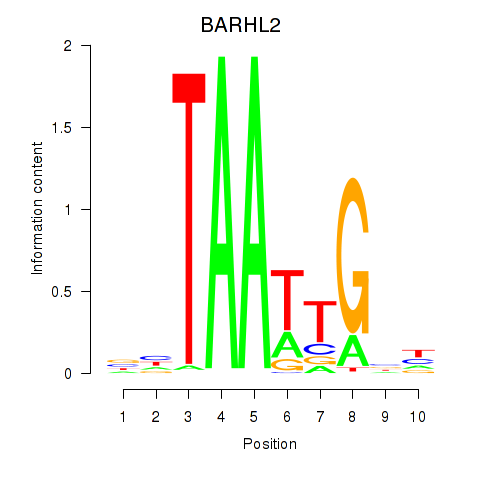
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_75351088 | 0.26 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr17_+_76037081 | 0.23 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr6_+_43968306 | 0.20 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr4_-_74853897 | 0.19 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr1_+_152943122 | 0.19 |
ENST00000328051.2
|
SPRR4
|
small proline-rich protein 4 |
| chr8_+_32579321 | 0.14 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr6_+_26199737 | 0.13 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr12_-_76879852 | 0.12 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr17_-_46799872 | 0.12 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr13_-_30160925 | 0.12 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr2_-_214017151 | 0.12 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr17_+_61151306 | 0.11 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr2_-_203735484 | 0.11 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr12_-_118628315 | 0.11 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chr5_+_142286887 | 0.10 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr11_+_34664014 | 0.10 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr19_-_7698599 | 0.10 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr14_-_91294472 | 0.10 |
ENST00000555975.1
|
CTD-3035D6.2
|
CTD-3035D6.2 |
| chr4_-_105416039 | 0.10 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr7_+_107224364 | 0.10 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr3_-_186262166 | 0.10 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr22_-_33968239 | 0.09 |
ENST00000452586.2
ENST00000421768.1 |
LARGE
|
like-glycosyltransferase |
| chr6_-_154751629 | 0.09 |
ENST00000424998.1
|
CNKSR3
|
CNKSR family member 3 |
| chr4_-_186570679 | 0.09 |
ENST00000451974.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr15_+_67841330 | 0.08 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr8_-_25281747 | 0.08 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr2_+_135596180 | 0.08 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_-_139726181 | 0.08 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr19_-_6690723 | 0.08 |
ENST00000601008.1
|
C3
|
complement component 3 |
| chr8_-_37411648 | 0.08 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr17_-_47045949 | 0.08 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr3_+_177159695 | 0.08 |
ENST00000442937.1
|
LINC00578
|
long intergenic non-protein coding RNA 578 |
| chr2_+_135596106 | 0.08 |
ENST00000356140.5
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr17_-_27188984 | 0.08 |
ENST00000582320.2
|
MIR144
|
microRNA 451b |
| chr2_+_47454054 | 0.07 |
ENST00000426892.1
|
AC106869.2
|
AC106869.2 |
| chr8_-_23712312 | 0.07 |
ENST00000290271.2
|
STC1
|
stanniocalcin 1 |
| chr12_+_27623565 | 0.07 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr12_-_86650077 | 0.07 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chrX_+_10126488 | 0.07 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_+_131958436 | 0.07 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr12_+_72061563 | 0.07 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr3_-_71777824 | 0.07 |
ENST00000469524.1
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr3_-_178984759 | 0.07 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr14_+_92789498 | 0.07 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr16_+_24621546 | 0.07 |
ENST00000566108.1
|
CTD-2540M10.1
|
CTD-2540M10.1 |
| chr8_-_116504448 | 0.07 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr15_+_94899183 | 0.06 |
ENST00000557742.1
|
MCTP2
|
multiple C2 domains, transmembrane 2 |
| chr15_+_77713299 | 0.06 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr5_-_146781153 | 0.06 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_180397256 | 0.06 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr2_-_208031943 | 0.06 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr4_-_170897045 | 0.06 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr4_+_70894130 | 0.06 |
ENST00000526767.1
ENST00000530128.1 ENST00000381057.3 |
HTN3
|
histatin 3 |
| chr11_-_795400 | 0.06 |
ENST00000526152.1
ENST00000456706.2 ENST00000528936.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr17_-_39123144 | 0.06 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr1_-_152386732 | 0.06 |
ENST00000271835.3
|
CRNN
|
cornulin |
| chr19_-_6057282 | 0.06 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr22_-_36357671 | 0.06 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr2_+_33359473 | 0.06 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr8_+_92114060 | 0.06 |
ENST00000518304.1
|
LRRC69
|
leucine rich repeat containing 69 |
| chr7_-_2883650 | 0.06 |
ENST00000544127.1
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr4_-_70653673 | 0.06 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr7_-_6066183 | 0.06 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr11_-_795286 | 0.06 |
ENST00000533385.1
ENST00000527723.1 |
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr17_-_38859996 | 0.06 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr7_-_141541221 | 0.06 |
ENST00000350549.3
ENST00000438520.1 |
PRSS37
|
protease, serine, 37 |
| chr6_-_10435032 | 0.06 |
ENST00000491317.1
ENST00000496285.1 ENST00000479822.1 ENST00000487130.1 |
LINC00518
|
long intergenic non-protein coding RNA 518 |
| chr12_+_64798826 | 0.05 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr4_-_122148620 | 0.05 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr1_+_2066252 | 0.05 |
ENST00000466352.1
|
PRKCZ
|
protein kinase C, zeta |
| chr20_-_45530365 | 0.05 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr11_-_33913708 | 0.05 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr12_+_104697504 | 0.05 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr9_+_6215799 | 0.05 |
ENST00000417746.2
ENST00000456383.2 |
IL33
|
interleukin 33 |
| chr8_+_94752349 | 0.05 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr12_-_52828147 | 0.05 |
ENST00000252245.5
|
KRT75
|
keratin 75 |
| chr1_+_92545862 | 0.05 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr12_-_91576561 | 0.05 |
ENST00000547568.2
ENST00000552962.1 |
DCN
|
decorin |
| chr13_-_103426081 | 0.05 |
ENST00000376022.1
ENST00000376021.4 |
TEX30
|
testis expressed 30 |
| chr6_+_43603552 | 0.05 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr6_+_170863353 | 0.05 |
ENST00000421512.1
|
TBP
|
TATA box binding protein |
| chr5_-_20575959 | 0.05 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr4_-_186732892 | 0.05 |
ENST00000451958.1
ENST00000439914.1 ENST00000428330.1 ENST00000429056.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_-_2883928 | 0.05 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr10_-_49813090 | 0.05 |
ENST00000249601.4
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr5_-_115890554 | 0.05 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_-_137842536 | 0.05 |
ENST00000512039.1
|
RP11-138I17.1
|
RP11-138I17.1 |
| chr1_-_243326612 | 0.05 |
ENST00000492145.1
ENST00000490813.1 ENST00000464936.1 |
CEP170
|
centrosomal protein 170kDa |
| chr11_+_34663913 | 0.05 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr2_-_114647327 | 0.05 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr10_+_69865866 | 0.05 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr2_+_86333340 | 0.05 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr7_-_14026063 | 0.05 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr4_-_76957214 | 0.05 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr16_+_8806800 | 0.05 |
ENST00000561870.1
ENST00000396600.2 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr12_+_52056548 | 0.05 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr1_+_84767289 | 0.05 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr15_+_64680003 | 0.05 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr5_-_78281775 | 0.05 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr4_+_169013666 | 0.05 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr2_-_231989808 | 0.05 |
ENST00000258400.3
|
HTR2B
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr1_-_205649580 | 0.05 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr5_-_127674883 | 0.04 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr5_-_133510456 | 0.04 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr16_+_30406423 | 0.04 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr5_+_140855495 | 0.04 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr5_-_13944652 | 0.04 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr5_-_78281603 | 0.04 |
ENST00000264914.4
|
ARSB
|
arylsulfatase B |
| chr16_-_80926457 | 0.04 |
ENST00000563626.1
ENST00000562231.1 |
RP11-314O13.1
|
RP11-314O13.1 |
| chr6_+_13182751 | 0.04 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr7_-_14029283 | 0.04 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr15_-_60683326 | 0.04 |
ENST00000559350.1
ENST00000558986.1 ENST00000560389.1 |
ANXA2
|
annexin A2 |
| chr16_+_8807419 | 0.04 |
ENST00000565016.1
ENST00000567812.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr4_+_130017268 | 0.04 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr8_+_32579341 | 0.04 |
ENST00000519240.1
ENST00000539990.1 |
NRG1
|
neuregulin 1 |
| chr5_-_1799965 | 0.04 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr9_-_125590818 | 0.04 |
ENST00000259467.4
|
PDCL
|
phosducin-like |
| chr8_+_104383728 | 0.04 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr6_+_155538093 | 0.04 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_-_150473127 | 0.04 |
ENST00000521001.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr11_-_111649074 | 0.04 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr4_+_86749045 | 0.04 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_+_45986511 | 0.04 |
ENST00000458629.1
ENST00000457814.1 |
CXCR6
|
chemokine (C-X-C motif) receptor 6 |
| chr7_-_14029515 | 0.04 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr7_+_99717230 | 0.04 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr9_-_28670283 | 0.04 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr15_+_76629064 | 0.04 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr9_+_125027127 | 0.04 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr1_+_40506255 | 0.04 |
ENST00000421589.1
|
CAP1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr18_+_21529811 | 0.04 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr5_-_1799932 | 0.04 |
ENST00000382647.7
ENST00000505059.2 |
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr12_+_111374375 | 0.04 |
ENST00000553177.1
ENST00000548368.1 ENST00000331096.2 ENST00000547607.1 |
RP1-46F2.2
|
RP1-46F2.2 |
| chr5_+_140593509 | 0.04 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr14_-_91720224 | 0.04 |
ENST00000238699.3
ENST00000531499.2 |
GPR68
|
G protein-coupled receptor 68 |
| chr18_+_55888767 | 0.04 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr4_+_41258786 | 0.04 |
ENST00000503431.1
ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr1_-_224624730 | 0.04 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr15_-_37393406 | 0.04 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr11_+_101983176 | 0.04 |
ENST00000524575.1
|
YAP1
|
Yes-associated protein 1 |
| chr6_+_130339710 | 0.04 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr6_+_161123270 | 0.04 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr2_-_203735976 | 0.04 |
ENST00000435143.1
|
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr18_+_20714525 | 0.04 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr1_+_222910538 | 0.04 |
ENST00000434700.1
ENST00000445590.2 |
FAM177B
|
family with sequence similarity 177, member B |
| chr14_+_67831576 | 0.04 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr5_+_150639360 | 0.04 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr2_+_143635067 | 0.04 |
ENST00000264170.4
|
KYNU
|
kynureninase |
| chrX_+_77154935 | 0.04 |
ENST00000481445.1
|
COX7B
|
cytochrome c oxidase subunit VIIb |
| chr18_-_46784778 | 0.04 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr3_+_32859510 | 0.04 |
ENST00000383763.5
|
TRIM71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr19_+_36606354 | 0.04 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr4_+_147096837 | 0.04 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_173020056 | 0.04 |
ENST00000239468.2
ENST00000404377.3 |
TNFSF18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr11_-_18211042 | 0.04 |
ENST00000527671.1
|
RP11-113D6.6
|
Uncharacterized protein |
| chr8_+_79503458 | 0.04 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr12_+_76653682 | 0.04 |
ENST00000553247.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chrX_+_108779870 | 0.04 |
ENST00000372107.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chrX_-_71351678 | 0.03 |
ENST00000609883.1
ENST00000545866.1 |
RGAG4
|
retrotransposon gag domain containing 4 |
| chr10_-_33405600 | 0.03 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr12_-_95510743 | 0.03 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr3_+_173116225 | 0.03 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr12_+_122326662 | 0.03 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr2_-_238322800 | 0.03 |
ENST00000392004.3
ENST00000433762.1 ENST00000347401.3 ENST00000353578.4 ENST00000346358.4 ENST00000392003.2 |
COL6A3
|
collagen, type VI, alpha 3 |
| chrX_+_107334983 | 0.03 |
ENST00000457035.1
ENST00000545696.1 |
ATG4A
|
autophagy related 4A, cysteine peptidase |
| chr8_+_11666649 | 0.03 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr18_-_33702078 | 0.03 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr7_-_99716940 | 0.03 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_106546808 | 0.03 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr14_-_23426231 | 0.03 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr7_-_130080977 | 0.03 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr1_+_224301787 | 0.03 |
ENST00000366862.5
ENST00000424254.2 |
FBXO28
|
F-box protein 28 |
| chr12_+_7014126 | 0.03 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr14_+_23709555 | 0.03 |
ENST00000430154.2
|
C14orf164
|
chromosome 14 open reading frame 164 |
| chr15_+_77712993 | 0.03 |
ENST00000336216.4
ENST00000381714.3 ENST00000558651.1 |
HMG20A
|
high mobility group 20A |
| chr16_-_69418553 | 0.03 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr16_-_46655538 | 0.03 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chrX_+_100224676 | 0.03 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr7_+_141408153 | 0.03 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr2_+_166095898 | 0.03 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr21_-_19858196 | 0.03 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr13_+_50656307 | 0.03 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr13_+_78315348 | 0.03 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr15_-_55657428 | 0.03 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr7_+_150065278 | 0.03 |
ENST00000519397.1
ENST00000479668.1 ENST00000540729.1 |
REPIN1
|
replication initiator 1 |
| chr9_-_133769225 | 0.03 |
ENST00000343079.1
|
QRFP
|
pyroglutamylated RFamide peptide |
| chr6_+_131571535 | 0.03 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr6_+_28249299 | 0.03 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr4_+_76439095 | 0.03 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr7_-_111032971 | 0.03 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr15_-_42264702 | 0.03 |
ENST00000220325.4
|
EHD4
|
EH-domain containing 4 |
| chr7_+_34697973 | 0.03 |
ENST00000381539.3
|
NPSR1
|
neuropeptide S receptor 1 |
| chrX_+_86772787 | 0.03 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr6_-_76203345 | 0.03 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_71384300 | 0.03 |
ENST00000504451.1
|
AMTN
|
amelotin |
| chr13_+_111855399 | 0.03 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr3_+_167453493 | 0.03 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr4_+_144312659 | 0.03 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_-_121379739 | 0.03 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr12_-_57328187 | 0.03 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr15_-_44116873 | 0.03 |
ENST00000267812.3
|
MFAP1
|
microfibrillar-associated protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.0 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 0.0 | 0.1 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.0 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.0 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0035038 | female pronucleus assembly(GO:0035038) |
| 0.0 | 0.1 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.0 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.0 | GO:0044035 | multi-organism catabolic process(GO:0044035) development of symbiont involved in interaction with host(GO:0044115) modulation of development of symbiont involved in interaction with host(GO:0044145) negative regulation of development of symbiont involved in interaction with host(GO:0044147) metabolism of substance in other organism involved in symbiotic interaction(GO:0052214) catabolism of substance in other organism involved in symbiotic interaction(GO:0052227) metabolism of macromolecule in other organism involved in symbiotic interaction(GO:0052229) catabolism by host of symbiont macromolecule(GO:0052360) catabolism by organism of macromolecule in other organism involved in symbiotic interaction(GO:0052361) catabolism by host of symbiont protein(GO:0052362) catabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052363) catabolism by host of substance in symbiont(GO:0052364) metabolism by host of symbiont macromolecule(GO:0052416) metabolism by host of symbiont protein(GO:0052417) metabolism by organism of protein in other organism involved in symbiotic interaction(GO:0052418) metabolism by host of substance in symbiont(GO:0052419) |
| 0.0 | 0.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.0 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |