Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
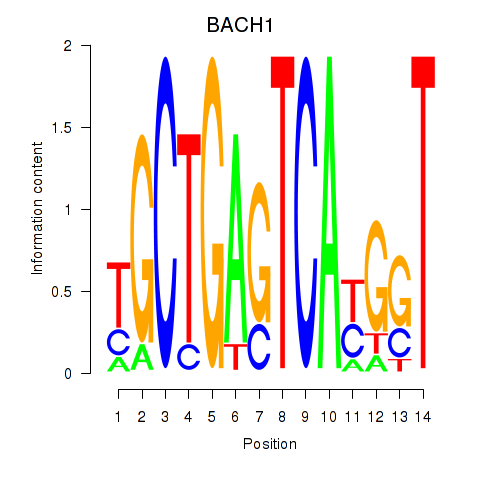
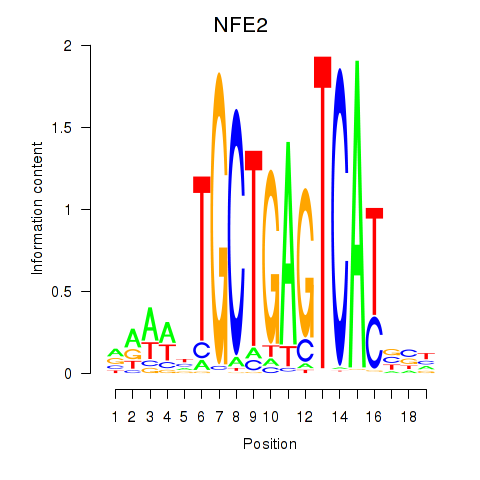
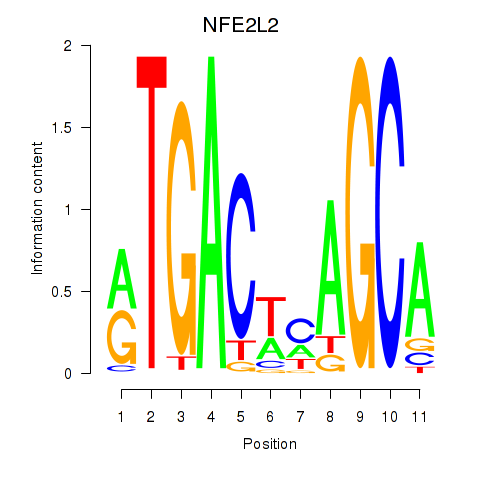
Results for BACH1_NFE2_NFE2L2
Z-value: 1.13
Transcription factors associated with BACH1_NFE2_NFE2L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BACH1
|
ENSG00000156273.11 | BTB domain and CNC homolog 1 |
|
NFE2
|
ENSG00000123405.9 | nuclear factor, erythroid 2 |
|
NFE2L2
|
ENSG00000116044.11 | nuclear factor, erythroid 2 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L2 | hg19_v2_chr2_-_178129551_178129572 | -0.84 | 3.6e-02 | Click! |
| BACH1 | hg19_v2_chr21_+_30672433_30672464 | -0.56 | 2.5e-01 | Click! |
| NFE2 | hg19_v2_chr12_-_54691668_54691725 | -0.42 | 4.1e-01 | Click! |
Activity profile of BACH1_NFE2_NFE2L2 motif
Sorted Z-values of BACH1_NFE2_NFE2L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22766766 | 4.49 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr20_+_44637526 | 1.48 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr21_+_42742429 | 1.32 |
ENST00000418103.1
|
MX2
|
myxovirus (influenza virus) resistance 2 (mouse) |
| chr12_-_122238464 | 0.87 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr7_-_127671674 | 0.78 |
ENST00000478726.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr16_+_89988259 | 0.78 |
ENST00000554444.1
ENST00000556565.1 |
TUBB3
|
Tubulin beta-3 chain |
| chr16_+_47496023 | 0.60 |
ENST00000567200.1
|
PHKB
|
phosphorylase kinase, beta |
| chr10_-_6019552 | 0.57 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr16_+_30077055 | 0.55 |
ENST00000564595.2
ENST00000569798.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_113763031 | 0.54 |
ENST00000259211.6
|
IL36A
|
interleukin 36, alpha |
| chr16_-_1429010 | 0.52 |
ENST00000513783.1
|
UNKL
|
unkempt family zinc finger-like |
| chr1_-_109968973 | 0.52 |
ENST00000271308.4
ENST00000538610.1 |
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr11_-_2924970 | 0.52 |
ENST00000533594.1
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr8_+_125283924 | 0.47 |
ENST00000517482.1
|
RP11-383J24.1
|
RP11-383J24.1 |
| chr17_-_38859996 | 0.47 |
ENST00000264651.2
|
KRT24
|
keratin 24 |
| chr22_-_19466454 | 0.44 |
ENST00000494054.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr3_+_14474178 | 0.43 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr1_-_200992827 | 0.42 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr14_+_103589789 | 0.40 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr14_+_35761540 | 0.39 |
ENST00000261479.4
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr4_+_22999152 | 0.38 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr16_+_30077098 | 0.37 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr22_-_19466643 | 0.36 |
ENST00000474226.1
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr2_-_220117867 | 0.35 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr6_-_3157760 | 0.35 |
ENST00000333628.3
|
TUBB2A
|
tubulin, beta 2A class IIa |
| chr14_-_57197224 | 0.33 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr1_+_96208646 | 0.33 |
ENST00000603401.1
|
RP11-286B14.2
|
RP11-286B14.2 |
| chr14_+_35761580 | 0.33 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr14_-_24732403 | 0.33 |
ENST00000206765.6
|
TGM1
|
transglutaminase 1 |
| chr10_-_6019984 | 0.33 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr22_-_19466683 | 0.32 |
ENST00000399523.1
ENST00000421968.2 ENST00000447868.1 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr11_-_102668879 | 0.32 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr3_+_46204959 | 0.31 |
ENST00000357422.2
|
CCR3
|
chemokine (C-C motif) receptor 3 |
| chr12_+_64846129 | 0.30 |
ENST00000540417.1
ENST00000539810.1 |
TBK1
|
TANK-binding kinase 1 |
| chr8_-_82395461 | 0.29 |
ENST00000256104.4
|
FABP4
|
fatty acid binding protein 4, adipocyte |
| chr11_+_72983246 | 0.29 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_-_102826434 | 0.28 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr16_-_1429627 | 0.27 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr22_-_39636914 | 0.27 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr14_-_24732368 | 0.27 |
ENST00000544573.1
|
TGM1
|
transglutaminase 1 |
| chr1_+_165796753 | 0.27 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr8_-_95220775 | 0.27 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr22_-_19466732 | 0.26 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr12_+_64845864 | 0.26 |
ENST00000538890.1
|
TBK1
|
TANK-binding kinase 1 |
| chr19_-_6670128 | 0.25 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr1_+_218683438 | 0.25 |
ENST00000443836.1
|
C1orf143
|
chromosome 1 open reading frame 143 |
| chr17_-_39769005 | 0.24 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr12_+_56075330 | 0.24 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr16_+_74330673 | 0.23 |
ENST00000219313.4
ENST00000540379.1 ENST00000567958.1 ENST00000568615.2 |
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr4_-_101111615 | 0.23 |
ENST00000273990.2
|
DDIT4L
|
DNA-damage-inducible transcript 4-like |
| chr6_+_44214824 | 0.23 |
ENST00000371646.5
ENST00000353801.3 |
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr4_+_79567057 | 0.22 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr9_+_140083099 | 0.22 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr14_-_24732738 | 0.22 |
ENST00000558074.1
ENST00000560226.1 |
TGM1
|
transglutaminase 1 |
| chr3_+_184016986 | 0.21 |
ENST00000417952.1
|
PSMD2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr18_+_68002675 | 0.21 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr11_+_10477733 | 0.20 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr10_+_91087651 | 0.20 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr21_+_33671264 | 0.20 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr5_+_35852797 | 0.19 |
ENST00000508941.1
|
IL7R
|
interleukin 7 receptor |
| chr12_+_10365404 | 0.19 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr4_-_76944621 | 0.19 |
ENST00000306602.1
|
CXCL10
|
chemokine (C-X-C motif) ligand 10 |
| chr5_-_90610200 | 0.19 |
ENST00000511918.1
ENST00000513626.1 ENST00000607854.1 |
LUCAT1
RP11-213H15.4
|
lung cancer associated transcript 1 (non-protein coding) RP11-213H15.4 |
| chr10_-_71892555 | 0.19 |
ENST00000307864.1
|
AIFM2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr19_+_56652643 | 0.18 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444 |
| chr14_-_23504087 | 0.18 |
ENST00000493471.2
ENST00000460922.2 |
PSMB5
|
proteasome (prosome, macropain) subunit, beta type, 5 |
| chr10_+_69865866 | 0.18 |
ENST00000354393.2
|
MYPN
|
myopalladin |
| chr11_-_67141640 | 0.18 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr7_+_139528952 | 0.17 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr7_+_42971799 | 0.17 |
ENST00000223324.2
|
MRPL32
|
mitochondrial ribosomal protein L32 |
| chr2_+_90248739 | 0.17 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr7_+_1084206 | 0.17 |
ENST00000444847.1
|
GPR146
|
G protein-coupled receptor 146 |
| chr19_+_50706866 | 0.17 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr6_+_44215603 | 0.17 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr22_+_19466980 | 0.17 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr2_+_201987200 | 0.17 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr5_-_93954227 | 0.17 |
ENST00000513200.3
ENST00000329378.7 ENST00000312498.7 |
KIAA0825
|
KIAA0825 |
| chr22_+_39353527 | 0.17 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr12_+_10366223 | 0.17 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr12_+_10365082 | 0.17 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_47447970 | 0.17 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr11_-_4599050 | 0.16 |
ENST00000307616.1
|
C11orf40
|
chromosome 11 open reading frame 40 |
| chr1_+_214163033 | 0.16 |
ENST00000607425.1
|
PROX1
|
prospero homeobox 1 |
| chr12_-_10826612 | 0.16 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr6_+_33378517 | 0.16 |
ENST00000428274.1
|
PHF1
|
PHD finger protein 1 |
| chr12_-_49523896 | 0.16 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr14_-_74462922 | 0.16 |
ENST00000553284.1
|
ENTPD5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr16_+_22308717 | 0.16 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr1_-_158656488 | 0.15 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr4_-_87374330 | 0.15 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr20_-_1306391 | 0.15 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr14_-_65769392 | 0.15 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr11_-_2924720 | 0.15 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr1_+_14075903 | 0.15 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr17_+_16284399 | 0.15 |
ENST00000535788.1
|
UBB
|
ubiquitin B |
| chr3_+_183903811 | 0.14 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr3_-_186262166 | 0.14 |
ENST00000307944.5
|
CRYGS
|
crystallin, gamma S |
| chr11_+_123396307 | 0.14 |
ENST00000456860.2
|
GRAMD1B
|
GRAM domain containing 1B |
| chr19_-_54604083 | 0.14 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr11_+_123325106 | 0.14 |
ENST00000525757.1
|
LINC01059
|
long intergenic non-protein coding RNA 1059 |
| chr2_-_191885686 | 0.14 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr22_-_39637135 | 0.14 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr21_+_33031935 | 0.14 |
ENST00000270142.6
ENST00000389995.4 |
SOD1
|
superoxide dismutase 1, soluble |
| chr3_+_184038234 | 0.14 |
ENST00000427607.1
ENST00000457456.1 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr20_+_42839600 | 0.13 |
ENST00000439943.1
ENST00000437730.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr18_+_61637159 | 0.13 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr2_+_54342533 | 0.13 |
ENST00000406041.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_11606275 | 0.13 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr7_+_139529040 | 0.13 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr6_-_11232891 | 0.13 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr17_+_18625336 | 0.13 |
ENST00000395671.4
ENST00000571542.1 ENST00000395672.2 ENST00000414850.2 ENST00000424146.2 |
TRIM16L
|
tripartite motif containing 16-like |
| chr11_-_128894053 | 0.13 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr3_+_184038073 | 0.13 |
ENST00000428387.1
ENST00000434061.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr3_+_184037466 | 0.13 |
ENST00000441154.1
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr15_-_34502197 | 0.12 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr19_-_10613421 | 0.12 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr22_+_21987005 | 0.12 |
ENST00000607942.1
ENST00000425975.1 ENST00000292779.3 |
CCDC116
|
coiled-coil domain containing 116 |
| chr17_+_30771279 | 0.12 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr18_-_55253830 | 0.12 |
ENST00000591215.1
|
FECH
|
ferrochelatase |
| chrX_-_24665353 | 0.12 |
ENST00000379144.2
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr12_-_57081940 | 0.12 |
ENST00000436399.2
|
PTGES3
|
prostaglandin E synthase 3 (cytosolic) |
| chr2_-_220119280 | 0.12 |
ENST00000392088.2
|
TUBA4A
|
tubulin, alpha 4a |
| chr16_-_1429674 | 0.12 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr16_-_30125177 | 0.12 |
ENST00000406256.3
|
GDPD3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr7_+_141463897 | 0.12 |
ENST00000247879.2
|
TAS2R3
|
taste receptor, type 2, member 3 |
| chr22_+_20008678 | 0.12 |
ENST00000434168.1
|
TANGO2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr4_-_44450814 | 0.12 |
ENST00000360029.3
|
KCTD8
|
potassium channel tetramerization domain containing 8 |
| chr15_-_43212836 | 0.12 |
ENST00000566931.1
ENST00000564431.1 ENST00000567274.1 |
TTBK2
|
tau tubulin kinase 2 |
| chr22_+_39916558 | 0.12 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr6_-_35888905 | 0.12 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr17_-_79604075 | 0.12 |
ENST00000374747.5
ENST00000539314.1 ENST00000331134.6 |
NPLOC4
|
nuclear protein localization 4 homolog (S. cerevisiae) |
| chr1_+_36554470 | 0.12 |
ENST00000373178.4
|
ADPRHL2
|
ADP-ribosylhydrolase like 2 |
| chr19_+_38865398 | 0.12 |
ENST00000585598.1
ENST00000602911.1 ENST00000592561.1 |
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr16_-_28222797 | 0.11 |
ENST00000569951.1
ENST00000565698.1 |
XPO6
|
exportin 6 |
| chr12_+_6644443 | 0.11 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_-_125399573 | 0.11 |
ENST00000339647.5
|
UBC
|
ubiquitin C |
| chr12_-_51611477 | 0.11 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr14_+_71165292 | 0.11 |
ENST00000553682.1
|
RP6-65G23.1
|
RP6-65G23.1 |
| chr17_-_8301132 | 0.11 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr17_-_42276574 | 0.11 |
ENST00000589805.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr4_+_123747834 | 0.11 |
ENST00000264498.3
|
FGF2
|
fibroblast growth factor 2 (basic) |
| chr21_+_19273574 | 0.11 |
ENST00000400128.1
|
CHODL
|
chondrolectin |
| chr18_-_72920372 | 0.11 |
ENST00000581620.1
ENST00000582437.1 |
ZADH2
|
zinc binding alcohol dehydrogenase domain containing 2 |
| chr17_+_16284104 | 0.11 |
ENST00000577958.1
ENST00000302182.3 ENST00000577640.1 |
UBB
|
ubiquitin B |
| chr5_+_179247759 | 0.11 |
ENST00000389805.4
ENST00000504627.1 ENST00000402874.3 ENST00000510187.1 |
SQSTM1
|
sequestosome 1 |
| chr19_-_44324750 | 0.11 |
ENST00000594049.1
ENST00000414615.2 |
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr20_+_10199468 | 0.11 |
ENST00000254976.2
ENST00000304886.2 |
SNAP25
|
synaptosomal-associated protein, 25kDa |
| chr3_+_100053542 | 0.11 |
ENST00000394140.4
|
NIT2
|
nitrilase family, member 2 |
| chr17_-_73150629 | 0.11 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr20_-_1306351 | 0.10 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr20_-_1309809 | 0.10 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr15_-_43212996 | 0.10 |
ENST00000567840.1
|
TTBK2
|
tau tubulin kinase 2 |
| chr18_+_21594384 | 0.10 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr20_-_634000 | 0.10 |
ENST00000381962.3
|
SRXN1
|
sulfiredoxin 1 |
| chr9_+_26746951 | 0.10 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chrX_-_24665208 | 0.10 |
ENST00000356768.4
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr11_-_47447767 | 0.10 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr19_+_38865176 | 0.10 |
ENST00000215071.4
|
PSMD8
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 |
| chr2_+_173955327 | 0.10 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr14_-_95942173 | 0.09 |
ENST00000334258.5
ENST00000557275.1 ENST00000553340.1 |
SYNE3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr17_-_7518145 | 0.09 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr2_-_233641265 | 0.09 |
ENST00000438786.1
ENST00000409779.1 ENST00000233826.3 |
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr17_-_28619059 | 0.09 |
ENST00000580709.1
|
BLMH
|
bleomycin hydrolase |
| chr13_+_78109804 | 0.09 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr19_-_46142680 | 0.09 |
ENST00000245925.3
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_46142637 | 0.09 |
ENST00000590043.1
ENST00000589876.1 |
EML2
|
echinoderm microtubule associated protein like 2 |
| chr9_+_116917807 | 0.09 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr17_-_79818354 | 0.09 |
ENST00000576541.1
ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr20_+_2796948 | 0.09 |
ENST00000361033.1
ENST00000380585.1 |
TMEM239
|
transmembrane protein 239 |
| chr19_-_14992264 | 0.09 |
ENST00000327462.2
|
OR7A17
|
olfactory receptor, family 7, subfamily A, member 17 |
| chr9_-_35112376 | 0.09 |
ENST00000488109.2
|
FAM214B
|
family with sequence similarity 214, member B |
| chr19_-_51568324 | 0.09 |
ENST00000595547.1
ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13
|
kallikrein-related peptidase 13 |
| chr13_+_114567131 | 0.08 |
ENST00000608651.1
|
GAS6-AS2
|
GAS6 antisense RNA 2 (head to head) |
| chrX_+_30671476 | 0.08 |
ENST00000378946.3
ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK
|
glycerol kinase |
| chr9_-_140082983 | 0.08 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr3_+_126423045 | 0.08 |
ENST00000290913.3
ENST00000508789.1 |
CHCHD6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr2_-_11605966 | 0.08 |
ENST00000307236.4
ENST00000542100.1 ENST00000546212.1 |
E2F6
|
E2F transcription factor 6 |
| chr3_+_120315149 | 0.08 |
ENST00000184266.2
|
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr7_-_75988321 | 0.08 |
ENST00000307630.3
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
| chrX_+_99899180 | 0.08 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr18_+_55712915 | 0.08 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr7_+_139529085 | 0.08 |
ENST00000539806.1
|
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr17_-_1395954 | 0.08 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr6_-_24799117 | 0.08 |
ENST00000565469.1
|
RP3-369A17.5
|
chromosome 6 open reading frame 229 |
| chr9_-_34458531 | 0.08 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr10_+_122216316 | 0.08 |
ENST00000398250.1
ENST00000439221.1 ENST00000398248.1 |
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A |
| chr8_+_39972170 | 0.08 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr22_-_32058416 | 0.08 |
ENST00000439502.2
|
PISD
|
phosphatidylserine decarboxylase |
| chr1_+_26605618 | 0.08 |
ENST00000270792.5
|
SH3BGRL3
|
SH3 domain binding glutamic acid-rich protein like 3 |
| chr9_-_26947220 | 0.08 |
ENST00000520884.1
|
PLAA
|
phospholipase A2-activating protein |
| chr4_-_987217 | 0.08 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr5_+_179125368 | 0.08 |
ENST00000502296.1
ENST00000504734.1 ENST00000415618.2 |
CANX
|
calnexin |
| chr16_+_57844549 | 0.08 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr17_-_61905005 | 0.07 |
ENST00000584574.1
ENST00000585145.1 ENST00000427159.2 |
FTSJ3
|
FtsJ homolog 3 (E. coli) |
| chr2_-_10588630 | 0.07 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr20_+_42839722 | 0.07 |
ENST00000442383.1
ENST00000435163.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr10_+_127661942 | 0.07 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr6_+_53948328 | 0.07 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr16_+_56642489 | 0.07 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr6_-_31745085 | 0.07 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BACH1_NFE2_NFE2L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.2 | 1.5 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.4 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.8 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.4 | GO:1900241 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.3 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 1.3 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.5 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 0.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0061114 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) negative regulation of bile acid biosynthetic process(GO:0070858) acinar cell differentiation(GO:0090425) negative regulation of bile acid metabolic process(GO:1904252) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.0 | 0.1 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.7 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.3 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.4 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.2 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.2 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.1 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.0 | GO:0060168 | regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 1.3 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) sperm plasma membrane(GO:0097524) |
| 0.1 | 0.2 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.6 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 2.5 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.1 | 0.4 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.3 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.8 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.0 | 0.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0019828 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.1 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 1.8 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.7 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |