Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
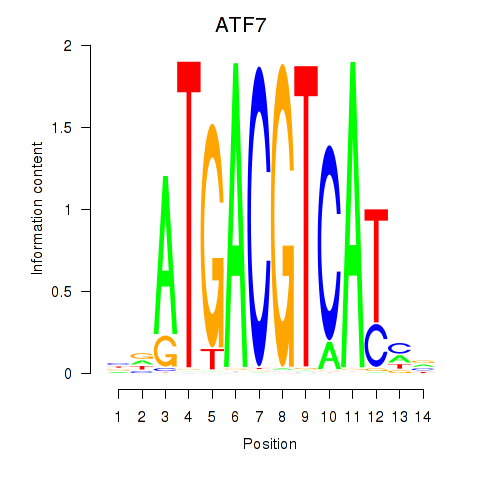
Results for ATF7
Z-value: 0.95
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | activating transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF7 | hg19_v2_chr12_-_54019755_54019804 | 0.77 | 7.5e-02 | Click! |
Activity profile of ATF7 motif
Sorted Z-values of ATF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_76829385 | 0.75 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr8_-_30013748 | 0.72 |
ENST00000607315.1
|
RP11-51J9.5
|
RP11-51J9.5 |
| chr18_+_76829441 | 0.70 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr17_-_43210580 | 0.63 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr2_+_171640291 | 0.61 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr7_-_65958525 | 0.59 |
ENST00000450353.1
ENST00000412091.1 |
GS1-124K5.3
|
GS1-124K5.3 |
| chr19_-_50979981 | 0.57 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chrX_+_48398053 | 0.54 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr6_-_27782548 | 0.53 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr22_-_44258360 | 0.51 |
ENST00000330884.4
ENST00000249130.5 |
SULT4A1
|
sulfotransferase family 4A, member 1 |
| chr14_-_77495007 | 0.51 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr5_-_175965008 | 0.50 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chrX_+_133941250 | 0.49 |
ENST00000445123.1
|
FAM122C
|
family with sequence similarity 122C |
| chr14_+_105219437 | 0.44 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr19_-_51014588 | 0.42 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr7_-_142247606 | 0.39 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr14_-_103989033 | 0.38 |
ENST00000553878.1
ENST00000557530.1 |
CKB
|
creatine kinase, brain |
| chr19_-_460996 | 0.37 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr19_-_5340730 | 0.34 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr19_+_49122548 | 0.33 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr17_+_8152590 | 0.33 |
ENST00000584044.1
ENST00000314666.6 ENST00000545834.1 ENST00000581242.1 |
PFAS
|
phosphoribosylformylglycinamidine synthase |
| chr19_+_46009837 | 0.32 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr6_+_139456226 | 0.32 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr7_-_45026159 | 0.32 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr17_+_73089382 | 0.31 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_+_153750622 | 0.31 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr2_-_220408260 | 0.30 |
ENST00000373891.2
|
CHPF
|
chondroitin polymerizing factor |
| chr6_+_126277842 | 0.30 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr9_-_125027079 | 0.29 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr2_+_219745020 | 0.29 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr13_-_101327028 | 0.29 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr1_+_85527987 | 0.28 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr1_-_228291136 | 0.28 |
ENST00000272139.4
|
C1orf35
|
chromosome 1 open reading frame 35 |
| chr14_+_61447832 | 0.28 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr16_+_81040794 | 0.26 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr4_-_2010562 | 0.26 |
ENST00000411649.1
ENST00000542778.1 ENST00000411638.2 ENST00000431323.1 |
NELFA
|
negative elongation factor complex member A |
| chr19_-_51014460 | 0.26 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chrX_-_135056106 | 0.26 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr13_+_49822041 | 0.25 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr5_+_112849373 | 0.25 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr3_+_158288999 | 0.25 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chrX_-_83757399 | 0.25 |
ENST00000373177.2
ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX
|
highly divergent homeobox |
| chr3_+_62304648 | 0.24 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr4_-_104119528 | 0.24 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chrX_-_135056216 | 0.24 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr19_-_47616992 | 0.23 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr15_+_91498089 | 0.23 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
| chr19_+_50979753 | 0.23 |
ENST00000597426.1
ENST00000334976.6 ENST00000376918.3 ENST00000598585.1 |
EMC10
|
ER membrane protein complex subunit 10 |
| chr9_-_34665983 | 0.23 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr3_-_119182506 | 0.23 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr11_-_82782952 | 0.22 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr17_+_4046964 | 0.22 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr11_-_65793948 | 0.22 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chr19_+_18043810 | 0.22 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr12_+_54378849 | 0.22 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr19_-_39402798 | 0.21 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr14_+_88851874 | 0.21 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr18_-_2571210 | 0.21 |
ENST00000577166.1
|
METTL4
|
methyltransferase like 4 |
| chr7_+_100464760 | 0.21 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr2_-_220408430 | 0.21 |
ENST00000243776.6
|
CHPF
|
chondroitin polymerizing factor |
| chr7_+_100210133 | 0.21 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr7_+_23221438 | 0.20 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr3_+_158288942 | 0.20 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr4_-_122744998 | 0.20 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr1_+_26496362 | 0.19 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr19_+_8455077 | 0.19 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr20_+_17949724 | 0.19 |
ENST00000377704.4
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chrX_-_47863348 | 0.19 |
ENST00000376943.3
ENST00000396965.1 ENST00000305127.6 |
ZNF182
|
zinc finger protein 182 |
| chr17_-_43209862 | 0.19 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr11_-_85376121 | 0.19 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr4_+_119512928 | 0.18 |
ENST00000567913.2
|
RP11-384K6.6
|
RP11-384K6.6 |
| chr7_-_124569864 | 0.18 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr5_+_139055055 | 0.18 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr6_+_150690028 | 0.18 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr11_+_102217936 | 0.18 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr3_+_158288960 | 0.18 |
ENST00000484955.1
ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr17_-_41322332 | 0.18 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr1_-_1284730 | 0.17 |
ENST00000378888.5
|
DVL1
|
dishevelled segment polarity protein 1 |
| chr7_+_5632436 | 0.17 |
ENST00000340250.6
ENST00000382361.3 |
FSCN1
|
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
| chr19_+_49122785 | 0.17 |
ENST00000598088.1
|
SPHK2
|
sphingosine kinase 2 |
| chr10_+_35484793 | 0.17 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr12_-_31881944 | 0.17 |
ENST00000537562.1
ENST00000537960.1 ENST00000536761.1 ENST00000542781.1 ENST00000457428.2 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr7_-_124569991 | 0.17 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr11_-_82782861 | 0.16 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr19_-_51014345 | 0.16 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr21_-_30365136 | 0.16 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr18_+_2571510 | 0.16 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr17_+_29421987 | 0.16 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr17_+_7155556 | 0.16 |
ENST00000570500.1
ENST00000574993.1 ENST00000396628.2 ENST00000573657.1 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr3_+_140660743 | 0.16 |
ENST00000453248.2
|
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr3_-_154042205 | 0.16 |
ENST00000329463.5
|
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr5_-_16509101 | 0.16 |
ENST00000399793.2
|
FAM134B
|
family with sequence similarity 134, member B |
| chr19_+_59055412 | 0.16 |
ENST00000593582.1
|
TRIM28
|
tripartite motif containing 28 |
| chr3_+_44803322 | 0.16 |
ENST00000481166.2
|
KIF15
|
kinesin family member 15 |
| chr11_-_62389577 | 0.15 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr1_-_115124257 | 0.15 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr17_+_43209967 | 0.15 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr8_-_146012660 | 0.15 |
ENST00000343459.4
ENST00000429371.2 ENST00000534445.1 |
ZNF34
|
zinc finger protein 34 |
| chr16_+_3068393 | 0.15 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr2_-_218706877 | 0.15 |
ENST00000446688.1
|
TNS1
|
tensin 1 |
| chr20_+_34203794 | 0.15 |
ENST00000374273.3
|
SPAG4
|
sperm associated antigen 4 |
| chr15_-_72766533 | 0.15 |
ENST00000562573.1
|
RP11-1007O24.3
|
RP11-1007O24.3 |
| chr19_+_36103631 | 0.15 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr3_+_140660634 | 0.15 |
ENST00000446041.2
ENST00000507429.1 ENST00000324194.6 |
SLC25A36
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36 |
| chr3_+_62304712 | 0.14 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr9_-_130477912 | 0.14 |
ENST00000543175.1
|
PTRH1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr10_+_24544249 | 0.14 |
ENST00000430453.2
|
KIAA1217
|
KIAA1217 |
| chr19_-_4902877 | 0.14 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr12_+_2921788 | 0.14 |
ENST00000228799.2
ENST00000419778.2 ENST00000542548.1 |
ITFG2
|
integrin alpha FG-GAP repeat containing 2 |
| chr2_-_190627481 | 0.14 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr11_-_62389449 | 0.14 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr17_+_74536164 | 0.13 |
ENST00000586148.1
|
PRCD
|
progressive rod-cone degeneration |
| chr9_+_131644398 | 0.13 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr2_-_114514181 | 0.13 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr5_+_131892603 | 0.13 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr16_+_19079311 | 0.13 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr18_-_2571437 | 0.13 |
ENST00000574676.1
ENST00000574538.1 ENST00000319888.6 |
METTL4
|
methyltransferase like 4 |
| chr6_+_150690133 | 0.13 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr19_+_10362882 | 0.13 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr3_-_107941209 | 0.13 |
ENST00000492106.1
|
IFT57
|
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr6_-_30523865 | 0.13 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr8_+_98788057 | 0.12 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr8_-_10697281 | 0.12 |
ENST00000524114.1
ENST00000553390.1 ENST00000554914.1 |
PINX1
SOX7
SOX7
|
PIN2/TERF1 interacting, telomerase inhibitor 1 SRY (sex determining region Y)-box 7 Transcription factor SOX-7; Uncharacterized protein; cDNA FLJ58508, highly similar to Transcription factor SOX-7 |
| chr7_+_100209979 | 0.12 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr6_-_106773291 | 0.12 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr10_-_105156198 | 0.12 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr19_+_52848659 | 0.12 |
ENST00000327920.8
|
ZNF610
|
zinc finger protein 610 |
| chr11_+_28129795 | 0.12 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr14_+_23025534 | 0.12 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr14_+_61447927 | 0.12 |
ENST00000451406.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr9_+_131644388 | 0.12 |
ENST00000372600.4
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr19_+_57901326 | 0.12 |
ENST00000596400.1
|
AC003002.6
|
Uncharacterized protein |
| chr5_-_178054014 | 0.12 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr11_-_77185094 | 0.12 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr11_+_12695944 | 0.12 |
ENST00000361905.4
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr3_-_154042235 | 0.11 |
ENST00000308361.6
ENST00000496811.1 ENST00000544526.1 |
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr19_+_57901208 | 0.11 |
ENST00000366197.5
ENST00000596282.1 ENST00000597400.1 ENST00000598895.1 ENST00000336128.7 ENST00000596617.1 |
ZNF548
AC003002.6
|
zinc finger protein 548 Uncharacterized protein |
| chr19_+_8455200 | 0.11 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr8_-_82754427 | 0.11 |
ENST00000353788.4
ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16
|
sorting nexin 16 |
| chr19_-_49122384 | 0.11 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr7_-_44924939 | 0.11 |
ENST00000395699.2
|
PURB
|
purine-rich element binding protein B |
| chr11_+_12696102 | 0.11 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr18_+_23806437 | 0.11 |
ENST00000578121.1
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr17_+_4046462 | 0.11 |
ENST00000577075.2
ENST00000575251.1 ENST00000301391.3 |
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr17_+_33914424 | 0.11 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr14_+_93651296 | 0.11 |
ENST00000283534.4
ENST00000557574.1 |
TMEM251
RP11-371E8.4
|
transmembrane protein 251 Uncharacterized protein |
| chr11_+_58938903 | 0.10 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr1_-_70671216 | 0.10 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr19_+_1941117 | 0.10 |
ENST00000255641.8
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr9_+_131644781 | 0.10 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr8_-_21966893 | 0.10 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr7_+_100209725 | 0.10 |
ENST00000223054.4
|
MOSPD3
|
motile sperm domain containing 3 |
| chr19_+_36486078 | 0.10 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chr6_+_56819895 | 0.10 |
ENST00000370748.3
|
BEND6
|
BEN domain containing 6 |
| chr14_+_96722152 | 0.10 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr9_-_131644202 | 0.10 |
ENST00000320665.6
ENST00000436267.2 |
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr19_+_36119929 | 0.10 |
ENST00000588161.1
ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42
|
RNA binding motif protein 42 |
| chr12_+_7013897 | 0.10 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr19_+_55996316 | 0.10 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr12_+_7014064 | 0.10 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr18_+_23806382 | 0.10 |
ENST00000400466.2
|
TAF4B
|
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
| chr14_+_96722539 | 0.09 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr10_-_27444143 | 0.09 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr20_+_42295745 | 0.09 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr1_+_228327943 | 0.09 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr4_+_1873155 | 0.09 |
ENST00000507820.1
ENST00000514045.1 |
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr3_+_44771088 | 0.09 |
ENST00000396048.2
|
ZNF501
|
zinc finger protein 501 |
| chr20_+_17949684 | 0.09 |
ENST00000377709.1
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr4_+_159131346 | 0.09 |
ENST00000508243.1
ENST00000296529.6 |
TMEM144
|
transmembrane protein 144 |
| chr5_-_178054105 | 0.09 |
ENST00000316308.4
|
CLK4
|
CDC-like kinase 4 |
| chr10_+_22634384 | 0.09 |
ENST00000376624.3
ENST00000376603.2 ENST00000376601.1 ENST00000538630.1 ENST00000456231.2 ENST00000313311.6 ENST00000435326.1 |
SPAG6
|
sperm associated antigen 6 |
| chr7_+_140396756 | 0.09 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr6_-_169654139 | 0.09 |
ENST00000366787.3
|
THBS2
|
thrombospondin 2 |
| chr1_-_153917700 | 0.09 |
ENST00000368646.2
|
DENND4B
|
DENN/MADD domain containing 4B |
| chr2_+_220408724 | 0.09 |
ENST00000421791.1
ENST00000373883.3 ENST00000451952.1 |
TMEM198
|
transmembrane protein 198 |
| chr4_-_104119488 | 0.09 |
ENST00000514974.1
|
CENPE
|
centromere protein E, 312kDa |
| chr6_-_106773610 | 0.09 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr5_-_175964366 | 0.09 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr4_+_78783674 | 0.08 |
ENST00000315567.8
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr17_+_41476327 | 0.08 |
ENST00000320033.4
|
ARL4D
|
ADP-ribosylation factor-like 4D |
| chr14_+_100485712 | 0.08 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr14_+_60716159 | 0.08 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr18_+_77867177 | 0.08 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr16_+_2563871 | 0.08 |
ENST00000330398.4
ENST00000568562.1 ENST00000569317.1 |
ATP6V0C
ATP6C
|
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c Uncharacterized protein |
| chr2_+_108994466 | 0.08 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr19_+_44100727 | 0.08 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr17_+_7155343 | 0.08 |
ENST00000573513.1
ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chrX_+_49832231 | 0.08 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr7_+_23221613 | 0.08 |
ENST00000410002.3
ENST00000413919.1 |
NUPL2
|
nucleoporin like 2 |
| chr6_-_32557610 | 0.08 |
ENST00000360004.5
|
HLA-DRB1
|
major histocompatibility complex, class II, DR beta 1 |
| chr9_-_131644306 | 0.08 |
ENST00000302586.3
|
CCBL1
|
cysteine conjugate-beta lyase, cytoplasmic |
| chr20_-_44420507 | 0.08 |
ENST00000243938.4
|
WFDC3
|
WAP four-disulfide core domain 3 |
| chr17_+_6918064 | 0.08 |
ENST00000546760.1
ENST00000552402.1 |
C17orf49
|
chromosome 17 open reading frame 49 |
| chr9_-_77643307 | 0.08 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr19_-_10341948 | 0.07 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr5_+_139055021 | 0.07 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr6_-_144416737 | 0.07 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr10_-_18940501 | 0.07 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr17_-_7155775 | 0.07 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chrX_-_7895755 | 0.07 |
ENST00000444736.1
ENST00000537427.1 ENST00000442940.1 |
PNPLA4
|
patatin-like phospholipase domain containing 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.3 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.4 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.2 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.2 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.5 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.3 | GO:0072642 | interferon-alpha secretion(GO:0072642) telomerase RNA stabilization(GO:0090669) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.3 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.8 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.3 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.1 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0010868 | negative regulation of triglyceride biosynthetic process(GO:0010868) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of muscle(GO:1901079) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.0 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.5 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.3 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |