Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
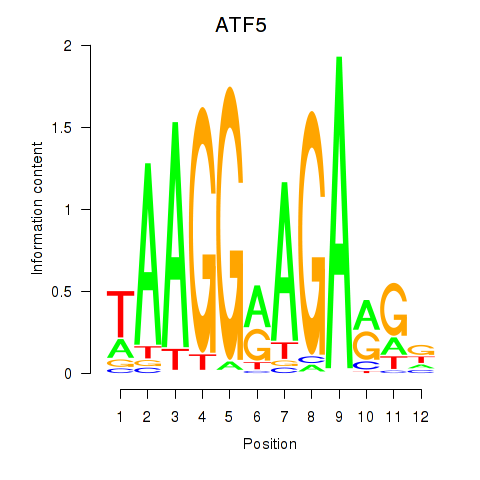
Results for ATF5
Z-value: 0.83
Transcription factors associated with ATF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF5
|
ENSG00000169136.4 | activating transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF5 | hg19_v2_chr19_+_50431959_50431998 | -0.52 | 2.9e-01 | Click! |
Activity profile of ATF5 motif
Sorted Z-values of ATF5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_52612705 | 0.74 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr14_+_32546145 | 0.63 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_-_188419078 | 0.41 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr19_+_42773371 | 0.38 |
ENST00000571942.2
|
CIC
|
capicua transcriptional repressor |
| chr14_+_32547434 | 0.37 |
ENST00000556191.1
ENST00000554090.1 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr18_-_19748379 | 0.36 |
ENST00000579431.1
|
GATA6-AS1
|
GATA6 antisense RNA 1 (head to head) |
| chr8_-_95449155 | 0.36 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr20_-_52612468 | 0.34 |
ENST00000422805.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chrX_-_19983260 | 0.34 |
ENST00000340625.3
|
CXorf23
|
chromosome X open reading frame 23 |
| chr14_+_32546274 | 0.31 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr2_-_114514181 | 0.27 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr3_-_101232019 | 0.26 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr3_+_119814070 | 0.26 |
ENST00000469070.1
|
RP11-18H7.1
|
RP11-18H7.1 |
| chr6_-_100016492 | 0.26 |
ENST00000369217.4
ENST00000369220.4 ENST00000482541.2 |
CCNC
|
cyclin C |
| chr1_+_61869748 | 0.26 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr8_+_95731904 | 0.26 |
ENST00000522422.1
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chr12_-_52967600 | 0.25 |
ENST00000549343.1
ENST00000305620.2 |
KRT74
|
keratin 74 |
| chr1_-_205053645 | 0.24 |
ENST00000367167.3
|
TMEM81
|
transmembrane protein 81 |
| chr2_-_188419200 | 0.23 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr1_+_223354486 | 0.22 |
ENST00000446145.1
|
RP11-239E10.3
|
RP11-239E10.3 |
| chr12_-_72057571 | 0.22 |
ENST00000548100.1
|
ZFC3H1
|
zinc finger, C3H1-type containing |
| chr16_-_72128270 | 0.21 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr6_-_88411911 | 0.21 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr12_+_32655048 | 0.21 |
ENST00000427716.2
ENST00000266482.3 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr9_+_19230433 | 0.21 |
ENST00000434457.2
ENST00000602925.1 |
DENND4C
|
DENN/MADD domain containing 4C |
| chrX_+_123095546 | 0.20 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chrX_-_44402231 | 0.20 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr6_+_87865262 | 0.20 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr11_-_117102768 | 0.20 |
ENST00000532301.1
|
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr6_-_100016527 | 0.19 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr3_-_183145873 | 0.19 |
ENST00000447025.2
ENST00000414362.2 ENST00000328913.3 |
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2 |
| chr14_+_50065459 | 0.19 |
ENST00000318317.4
|
LRR1
|
leucine rich repeat protein 1 |
| chr12_-_100656134 | 0.19 |
ENST00000548313.1
|
DEPDC4
|
DEP domain containing 4 |
| chr21_-_35773370 | 0.19 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr6_-_100016678 | 0.19 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr7_-_127672146 | 0.18 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr1_-_46598371 | 0.18 |
ENST00000372006.1
ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr4_-_84376953 | 0.18 |
ENST00000510985.1
ENST00000295488.3 |
HELQ
|
helicase, POLQ-like |
| chr12_-_72057638 | 0.18 |
ENST00000552037.1
ENST00000378743.3 |
ZFC3H1
|
zinc finger, C3H1-type containing |
| chrX_+_123095860 | 0.18 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr5_-_39219705 | 0.18 |
ENST00000351578.6
|
FYB
|
FYN binding protein |
| chr8_+_95732095 | 0.17 |
ENST00000414645.2
|
DPY19L4
|
dpy-19-like 4 (C. elegans) |
| chrX_+_123095890 | 0.16 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr3_-_88108192 | 0.16 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr1_+_101361626 | 0.16 |
ENST00000370112.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr6_+_76599809 | 0.16 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr17_+_38337491 | 0.16 |
ENST00000538981.1
|
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr8_+_67344710 | 0.15 |
ENST00000379385.4
ENST00000396623.3 ENST00000415254.1 |
ADHFE1
|
alcohol dehydrogenase, iron containing, 1 |
| chr4_+_2800722 | 0.15 |
ENST00000508385.1
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr9_-_6007787 | 0.14 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr17_+_38219063 | 0.14 |
ENST00000584985.1
ENST00000264637.4 ENST00000450525.2 |
THRA
|
thyroid hormone receptor, alpha |
| chrX_+_123095155 | 0.14 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chr11_-_6462210 | 0.14 |
ENST00000265983.3
|
HPX
|
hemopexin |
| chrX_-_47489244 | 0.14 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr3_+_193853927 | 0.14 |
ENST00000232424.3
|
HES1
|
hes family bHLH transcription factor 1 |
| chr14_+_50065376 | 0.13 |
ENST00000298288.6
|
LRR1
|
leucine rich repeat protein 1 |
| chr2_+_173724771 | 0.13 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr6_+_114178512 | 0.13 |
ENST00000368635.4
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate |
| chr1_+_206137237 | 0.13 |
ENST00000468509.1
ENST00000367129.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr8_+_79428539 | 0.13 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr10_+_104178946 | 0.12 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr11_-_108408895 | 0.12 |
ENST00000443411.1
ENST00000533052.1 |
EXPH5
|
exophilin 5 |
| chr4_-_122085469 | 0.12 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr3_+_112930306 | 0.12 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr14_-_36988882 | 0.12 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr7_-_100061869 | 0.11 |
ENST00000332375.3
|
C7orf61
|
chromosome 7 open reading frame 61 |
| chr5_+_50678921 | 0.11 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr16_+_57438679 | 0.11 |
ENST00000219244.4
|
CCL17
|
chemokine (C-C motif) ligand 17 |
| chr2_-_87248975 | 0.11 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr5_+_110559312 | 0.11 |
ENST00000508074.1
|
CAMK4
|
calcium/calmodulin-dependent protein kinase IV |
| chr12_-_115121962 | 0.11 |
ENST00000349155.2
|
TBX3
|
T-box 3 |
| chr15_+_63481668 | 0.11 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr1_-_27930102 | 0.11 |
ENST00000247087.5
ENST00000374011.2 |
AHDC1
|
AT hook, DNA binding motif, containing 1 |
| chr4_+_84377115 | 0.11 |
ENST00000295491.4
ENST00000507019.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr19_+_41509851 | 0.11 |
ENST00000593831.1
ENST00000330446.5 |
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr17_+_44790515 | 0.10 |
ENST00000576346.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr16_-_72127456 | 0.10 |
ENST00000562153.1
|
TXNL4B
|
thioredoxin-like 4B |
| chr14_-_54420133 | 0.10 |
ENST00000559501.1
ENST00000558984.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr1_+_160321120 | 0.10 |
ENST00000424754.1
|
NCSTN
|
nicastrin |
| chr5_-_39219641 | 0.10 |
ENST00000509072.1
ENST00000504542.1 ENST00000505428.1 ENST00000506557.1 |
FYB
|
FYN binding protein |
| chr3_+_19988736 | 0.10 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr18_-_812517 | 0.10 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr11_+_27076764 | 0.10 |
ENST00000525090.1
|
BBOX1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr15_+_78556428 | 0.10 |
ENST00000394855.3
ENST00000489435.2 |
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr3_+_101504200 | 0.09 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr12_+_32655110 | 0.09 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr5_+_82767487 | 0.09 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr8_+_145294015 | 0.09 |
ENST00000544576.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr22_+_37678505 | 0.09 |
ENST00000402997.1
ENST00000405206.3 |
CYTH4
|
cytohesin 4 |
| chr18_-_53089723 | 0.09 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr2_+_217498105 | 0.09 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr16_+_31271274 | 0.08 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr8_+_38677850 | 0.08 |
ENST00000518809.1
ENST00000520611.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_111765562 | 0.08 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr18_-_812231 | 0.08 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr12_-_120884175 | 0.08 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr4_+_84377198 | 0.08 |
ENST00000507349.1
ENST00000509970.1 ENST00000505719.1 |
MRPS18C
|
mitochondrial ribosomal protein S18C |
| chr5_+_82767284 | 0.08 |
ENST00000265077.3
|
VCAN
|
versican |
| chr19_-_44285401 | 0.08 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr16_+_28943260 | 0.08 |
ENST00000538922.1
ENST00000324662.3 ENST00000567541.1 |
CD19
|
CD19 molecule |
| chr19_-_15090488 | 0.08 |
ENST00000594383.1
ENST00000598504.1 ENST00000597262.1 |
SLC1A6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chrX_-_13835461 | 0.08 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr16_-_57513657 | 0.07 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr10_-_7453445 | 0.07 |
ENST00000379713.3
ENST00000397167.1 ENST00000397160.3 |
SFMBT2
|
Scm-like with four mbt domains 2 |
| chr5_+_175223313 | 0.07 |
ENST00000359546.4
|
CPLX2
|
complexin 2 |
| chr19_-_38916839 | 0.07 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr3_-_182698381 | 0.07 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr3_+_19988885 | 0.07 |
ENST00000422242.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr2_+_191334212 | 0.07 |
ENST00000444317.1
ENST00000535751.1 |
MFSD6
|
major facilitator superfamily domain containing 6 |
| chr3_-_127441406 | 0.07 |
ENST00000487473.1
ENST00000484451.1 |
MGLL
|
monoglyceride lipase |
| chr12_-_54778444 | 0.07 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr12_+_120884222 | 0.07 |
ENST00000551765.1
ENST00000229384.5 |
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
| chr8_+_37553261 | 0.07 |
ENST00000331569.4
|
ZNF703
|
zinc finger protein 703 |
| chr6_-_31514333 | 0.07 |
ENST00000376151.4
|
ATP6V1G2
|
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 |
| chr17_+_41005283 | 0.07 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr3_+_19988566 | 0.07 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr2_-_225434538 | 0.07 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr7_-_115670792 | 0.07 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr2_-_214016314 | 0.06 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr16_-_30537839 | 0.06 |
ENST00000380412.5
|
ZNF768
|
zinc finger protein 768 |
| chr17_+_33474826 | 0.06 |
ENST00000268876.5
ENST00000433649.1 ENST00000378449.1 |
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr5_+_125967372 | 0.06 |
ENST00000357147.3
|
C5orf48
|
chromosome 5 open reading frame 48 |
| chr5_+_82767583 | 0.06 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr11_+_124735282 | 0.06 |
ENST00000397801.1
|
ROBO3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr22_-_26875345 | 0.06 |
ENST00000398141.1
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr8_+_100025476 | 0.06 |
ENST00000355155.1
ENST00000357162.2 ENST00000358544.2 ENST00000395996.1 ENST00000441350.2 |
VPS13B
|
vacuolar protein sorting 13 homolog B (yeast) |
| chr11_-_18956556 | 0.06 |
ENST00000302797.3
|
MRGPRX1
|
MAS-related GPR, member X1 |
| chr14_-_21567009 | 0.06 |
ENST00000556174.1
ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219
|
zinc finger protein 219 |
| chr2_-_197036289 | 0.05 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr12_+_104680659 | 0.05 |
ENST00000526691.1
ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1
|
thioredoxin reductase 1 |
| chr19_+_15160130 | 0.05 |
ENST00000427043.3
|
CASP14
|
caspase 14, apoptosis-related cysteine peptidase |
| chr17_+_55055466 | 0.05 |
ENST00000262288.3
ENST00000572710.1 ENST00000575395.1 |
SCPEP1
|
serine carboxypeptidase 1 |
| chrX_+_131157609 | 0.05 |
ENST00000496850.1
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr9_+_131902283 | 0.05 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chrX_-_122866874 | 0.05 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr14_-_39901618 | 0.05 |
ENST00000554932.1
ENST00000298097.7 |
FBXO33
|
F-box protein 33 |
| chr7_-_115670804 | 0.05 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr9_+_109625378 | 0.04 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr3_-_99569821 | 0.04 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr4_+_48807155 | 0.04 |
ENST00000504654.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr10_-_99531709 | 0.04 |
ENST00000266066.3
|
SFRP5
|
secreted frizzled-related protein 5 |
| chr14_+_96968707 | 0.04 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr2_+_189839046 | 0.04 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr11_-_33891362 | 0.04 |
ENST00000395833.3
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr22_-_26875631 | 0.04 |
ENST00000402105.3
|
HPS4
|
Hermansky-Pudlak syndrome 4 |
| chr2_-_209118974 | 0.04 |
ENST00000415913.1
ENST00000415282.1 ENST00000446179.1 |
IDH1
|
isocitrate dehydrogenase 1 (NADP+), soluble |
| chrX_+_133930798 | 0.04 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr9_+_2621798 | 0.04 |
ENST00000382100.3
|
VLDLR
|
very low density lipoprotein receptor |
| chr19_-_18366182 | 0.04 |
ENST00000355502.3
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific |
| chr15_-_90222610 | 0.04 |
ENST00000300055.5
|
PLIN1
|
perilipin 1 |
| chr14_-_21566731 | 0.04 |
ENST00000360947.3
|
ZNF219
|
zinc finger protein 219 |
| chr19_+_55795493 | 0.03 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr12_-_1920886 | 0.03 |
ENST00000536846.2
ENST00000538027.2 ENST00000538450.1 |
CACNA2D4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr19_-_10491234 | 0.03 |
ENST00000524462.1
ENST00000531836.1 ENST00000525621.1 |
TYK2
|
tyrosine kinase 2 |
| chr2_-_190927447 | 0.03 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr16_-_72127550 | 0.03 |
ENST00000268483.3
|
TXNL4B
|
thioredoxin-like 4B |
| chr1_-_22109484 | 0.03 |
ENST00000529637.1
|
USP48
|
ubiquitin specific peptidase 48 |
| chr1_+_206579736 | 0.03 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr12_-_24103954 | 0.03 |
ENST00000441133.2
ENST00000545921.1 |
SOX5
|
SRY (sex determining region Y)-box 5 |
| chr9_-_116065551 | 0.03 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr15_-_90222642 | 0.03 |
ENST00000430628.2
|
PLIN1
|
perilipin 1 |
| chrX_+_122318318 | 0.03 |
ENST00000371251.1
ENST00000371256.5 |
GRIA3
|
glutamate receptor, ionotropic, AMPA 3 |
| chr19_-_10491130 | 0.03 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr1_-_151804222 | 0.03 |
ENST00000392697.3
|
RORC
|
RAR-related orphan receptor C |
| chr5_+_137203465 | 0.03 |
ENST00000239926.4
|
MYOT
|
myotilin |
| chr5_+_34757309 | 0.03 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr5_+_158527630 | 0.03 |
ENST00000523301.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr17_+_33474860 | 0.03 |
ENST00000394570.2
|
UNC45B
|
unc-45 homolog B (C. elegans) |
| chr8_+_107738343 | 0.03 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr3_+_23958470 | 0.03 |
ENST00000434031.2
ENST00000413699.1 ENST00000456530.2 |
RPL15
|
ribosomal protein L15 |
| chr3_-_23958402 | 0.03 |
ENST00000415901.2
ENST00000416026.2 ENST00000412028.1 ENST00000388759.3 ENST00000437230.1 |
NKIRAS1
|
NFKB inhibitor interacting Ras-like 1 |
| chr8_+_107738240 | 0.02 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr1_+_36023370 | 0.02 |
ENST00000356090.4
ENST00000373243.2 |
NCDN
|
neurochondrin |
| chr12_-_12491608 | 0.02 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr1_-_11863571 | 0.02 |
ENST00000376583.3
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr17_-_46691990 | 0.02 |
ENST00000576562.1
|
HOXB8
|
homeobox B8 |
| chr11_+_57310114 | 0.02 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr14_+_79746249 | 0.02 |
ENST00000428277.2
|
NRXN3
|
neurexin 3 |
| chr5_-_132200477 | 0.02 |
ENST00000296875.2
|
GDF9
|
growth differentiation factor 9 |
| chr1_-_54872059 | 0.02 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr16_-_72206034 | 0.02 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr17_+_33914424 | 0.02 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr7_-_78400364 | 0.02 |
ENST00000535697.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_+_156887027 | 0.02 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr14_+_79745746 | 0.02 |
ENST00000281127.7
|
NRXN3
|
neurexin 3 |
| chr1_-_11863171 | 0.02 |
ENST00000376592.1
|
MTHFR
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr5_+_132009675 | 0.02 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr12_-_54778244 | 0.02 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr18_-_24128496 | 0.02 |
ENST00000417602.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr22_-_36236623 | 0.02 |
ENST00000405409.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr1_+_16083123 | 0.02 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_+_30075463 | 0.02 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr11_+_28724129 | 0.01 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr5_-_156390230 | 0.01 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr12_-_54778471 | 0.01 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_-_202776392 | 0.01 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr7_+_73498118 | 0.01 |
ENST00000336180.2
|
LIMK1
|
LIM domain kinase 1 |
| chr1_-_156460391 | 0.01 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr12_-_54652060 | 0.01 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chr15_+_77287426 | 0.01 |
ENST00000558012.1
ENST00000267939.5 ENST00000379595.3 |
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 0.2 | GO:0015993 | molecular hydrogen transport(GO:0015993) |
| 0.0 | 0.6 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 | 0.1 | GO:2000974 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.0 | 0.1 | GO:0002372 | myeloid dendritic cell cytokine production(GO:0002372) |
| 0.0 | 0.4 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.1 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.1 | GO:0017055 | female courtship behavior(GO:0008050) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0061151 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.1 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) thyroid hormone binding(GO:0070324) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |