Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
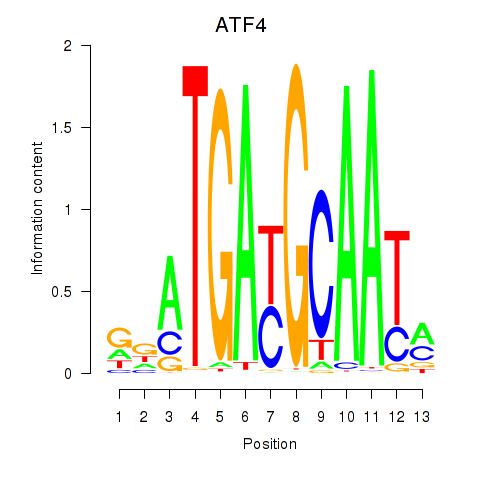
Results for ATF4
Z-value: 0.23
Transcription factors associated with ATF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF4
|
ENSG00000128272.10 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF4 | hg19_v2_chr22_+_39916558_39916579 | -0.78 | 6.7e-02 | Click! |
Activity profile of ATF4 motif
Sorted Z-values of ATF4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_41245160 | 0.20 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr4_-_70725856 | 0.13 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr17_-_40828969 | 0.12 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_+_73089382 | 0.12 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr17_-_40829026 | 0.09 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr19_-_51014588 | 0.08 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr17_+_39846114 | 0.08 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr1_+_239882842 | 0.08 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr5_+_174151536 | 0.08 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr17_+_1665306 | 0.08 |
ENST00000571360.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chrX_-_80457385 | 0.08 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr17_-_77967433 | 0.08 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_-_38412683 | 0.07 |
ENST00000373024.3
ENST00000373023.2 |
INPP5B
|
inositol polyphosphate-5-phosphatase, 75kDa |
| chr10_-_21806759 | 0.07 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr17_-_77813186 | 0.06 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr10_+_14880287 | 0.06 |
ENST00000437161.2
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr1_+_52521928 | 0.06 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr10_-_14880002 | 0.06 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr19_-_51014460 | 0.05 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr17_+_1665253 | 0.05 |
ENST00000254722.4
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_+_57849048 | 0.05 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr10_+_111765562 | 0.05 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr16_-_28550348 | 0.05 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr1_-_167883327 | 0.05 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chrX_+_48398053 | 0.05 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr16_-_28550320 | 0.04 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr12_+_57914742 | 0.04 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr9_-_34665983 | 0.04 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr11_+_58938903 | 0.04 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr7_-_99006443 | 0.04 |
ENST00000350498.3
|
PDAP1
|
PDGFA associated protein 1 |
| chr2_+_46769798 | 0.04 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr7_+_48211048 | 0.04 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr19_-_51014345 | 0.04 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr14_+_75894391 | 0.03 |
ENST00000419727.2
|
JDP2
|
Jun dimerization protein 2 |
| chr1_-_149832704 | 0.03 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr16_+_67694849 | 0.03 |
ENST00000602551.1
ENST00000458121.2 ENST00000219255.3 |
PARD6A
|
par-6 family cell polarity regulator alpha |
| chr14_+_24563262 | 0.03 |
ENST00000559250.1
ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr7_-_138386097 | 0.03 |
ENST00000421622.1
|
SVOPL
|
SVOP-like |
| chr19_-_4902877 | 0.03 |
ENST00000381781.2
|
ARRDC5
|
arrestin domain containing 5 |
| chr5_+_67586465 | 0.03 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr21_-_44495964 | 0.03 |
ENST00000398168.1
ENST00000398165.3 |
CBS
|
cystathionine-beta-synthase |
| chr5_+_44809027 | 0.03 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr1_-_204380919 | 0.02 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr15_+_40886439 | 0.02 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr10_+_95848824 | 0.02 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr10_-_113943447 | 0.02 |
ENST00000369425.1
ENST00000348367.4 ENST00000423155.1 |
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr4_-_153457197 | 0.02 |
ENST00000281708.4
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr19_-_47287990 | 0.02 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.2 |
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr15_+_40886199 | 0.02 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr16_-_67694597 | 0.02 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr1_+_162351503 | 0.02 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chr19_-_47288162 | 0.02 |
ENST00000594991.1
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr1_+_162601774 | 0.02 |
ENST00000415555.1
|
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr11_-_128737259 | 0.02 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr14_+_24563510 | 0.02 |
ENST00000545054.2
ENST00000561286.1 ENST00000558096.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr14_-_92413727 | 0.02 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr8_-_116681686 | 0.02 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr7_-_48068699 | 0.02 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr12_+_56862301 | 0.02 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr21_-_44495919 | 0.02 |
ENST00000398158.1
|
CBS
|
cystathionine-beta-synthase |
| chr22_-_43485381 | 0.02 |
ENST00000331018.7
ENST00000266254.7 ENST00000445824.1 |
TTLL1
|
tubulin tyrosine ligase-like family, member 1 |
| chr7_-_48068671 | 0.02 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr1_-_44497024 | 0.02 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_-_76836963 | 0.01 |
ENST00000312010.6
|
USP36
|
ubiquitin specific peptidase 36 |
| chr4_+_95129061 | 0.01 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_-_121944491 | 0.01 |
ENST00000331178.4
ENST00000427185.2 ENST00000442488.2 |
FEZF1
|
FEZ family zinc finger 1 |
| chr12_-_25101920 | 0.01 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr11_-_8680383 | 0.01 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr15_+_89182156 | 0.01 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr12_+_57623477 | 0.01 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_-_25102252 | 0.01 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_+_1665345 | 0.01 |
ENST00000576406.1
ENST00000571149.1 |
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr2_-_46769694 | 0.01 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr17_+_46133307 | 0.01 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr3_+_11314072 | 0.01 |
ENST00000444619.1
|
ATG7
|
autophagy related 7 |
| chr12_+_6493406 | 0.01 |
ENST00000543190.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr9_+_80912059 | 0.01 |
ENST00000347159.2
ENST00000376588.3 |
PSAT1
|
phosphoserine aminotransferase 1 |
| chr5_+_167956121 | 0.01 |
ENST00000338333.4
|
FBLL1
|
fibrillarin-like 1 |
| chr22_+_42229100 | 0.01 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr3_-_33686743 | 0.00 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr4_+_95128996 | 0.00 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr7_+_99006232 | 0.00 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr11_-_77185094 | 0.00 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_+_102106829 | 0.00 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr4_+_95128748 | 0.00 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_+_52521797 | 0.00 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr12_-_46662772 | 0.00 |
ENST00000549049.1
ENST00000439706.1 ENST00000398637.5 |
SLC38A1
|
solute carrier family 38, member 1 |
| chr11_+_62649158 | 0.00 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr11_-_128737163 | 0.00 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr5_+_143584814 | 0.00 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr14_-_100841794 | 0.00 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr7_+_99006550 | 0.00 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr14_-_88282595 | 0.00 |
ENST00000554519.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr1_-_216978709 | 0.00 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr22_+_50919995 | 0.00 |
ENST00000362068.2
ENST00000395737.1 |
ADM2
|
adrenomedullin 2 |
| chr6_+_73076432 | 0.00 |
ENST00000414192.2
|
RIMS1
|
regulating synaptic membrane exocytosis 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.0 | GO:0009093 | cysteine biosynthetic process from serine(GO:0006535) cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.0 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.0 | GO:0098809 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |