Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
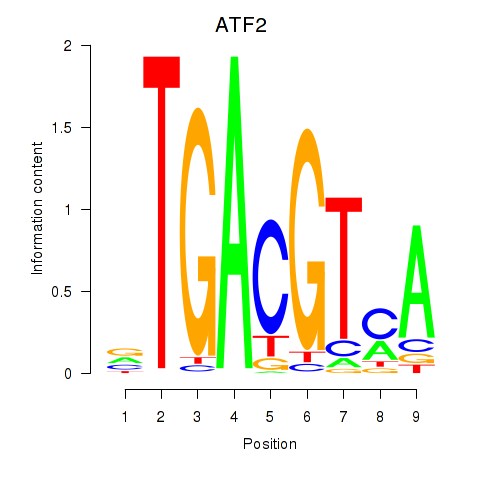
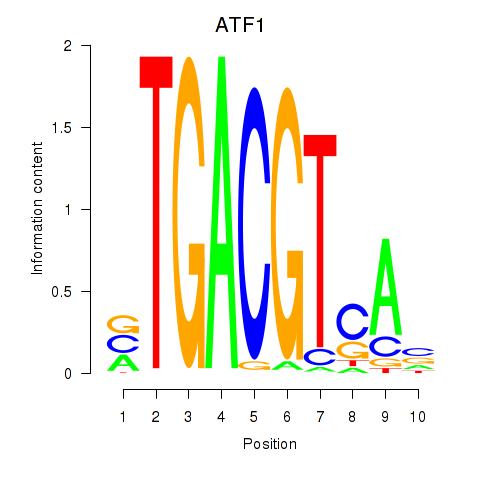
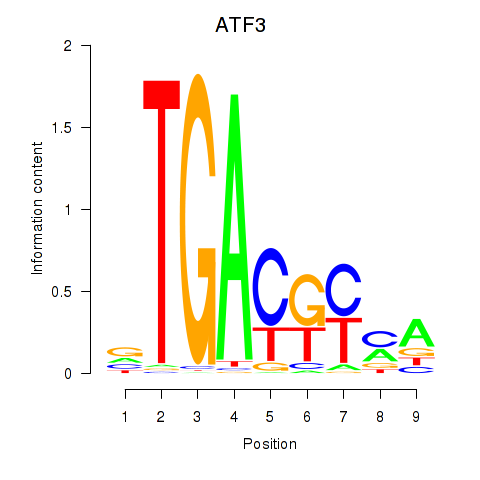
Results for ATF2_ATF1_ATF3
Z-value: 1.36
Transcription factors associated with ATF2_ATF1_ATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF2
|
ENSG00000115966.12 | activating transcription factor 2 |
|
ATF1
|
ENSG00000123268.4 | activating transcription factor 1 |
|
ATF3
|
ENSG00000162772.12 | activating transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF1 | hg19_v2_chr12_+_51158263_51158395 | 0.60 | 2.1e-01 | Click! |
| ATF2 | hg19_v2_chr2_-_176032843_176032941 | -0.11 | 8.4e-01 | Click! |
| ATF3 | hg19_v2_chr1_+_212782012_212782257 | -0.06 | 9.1e-01 | Click! |
Activity profile of ATF2_ATF1_ATF3 motif
Sorted Z-values of ATF2_ATF1_ATF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_103876528 | 2.34 |
ENST00000522939.1
ENST00000524007.1 |
KB-1507C5.2
|
HCG15011, isoform CRA_a; Protein LOC100996457 |
| chrX_-_135056106 | 1.26 |
ENST00000433339.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chrX_-_135056216 | 1.11 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr5_+_133707252 | 1.01 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr18_+_57567180 | 0.98 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr6_+_89791507 | 0.98 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr6_+_139456226 | 0.92 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr2_-_68290106 | 0.91 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr3_-_101395936 | 0.85 |
ENST00000461821.1
|
ZBTB11
|
zinc finger and BTB domain containing 11 |
| chr2_+_171640291 | 0.82 |
ENST00000409885.1
|
ERICH2
|
glutamate-rich 2 |
| chr4_-_113558014 | 0.74 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chr8_+_26240414 | 0.73 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr4_-_76598326 | 0.71 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr6_-_121655850 | 0.69 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr8_+_125486939 | 0.68 |
ENST00000303545.3
|
RNF139
|
ring finger protein 139 |
| chr15_-_32162833 | 0.68 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chr4_-_54930790 | 0.67 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr7_+_107220660 | 0.67 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr8_-_17104099 | 0.67 |
ENST00000524358.1
|
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr6_+_126277842 | 0.67 |
ENST00000229633.5
|
HINT3
|
histidine triad nucleotide binding protein 3 |
| chr4_-_68566832 | 0.66 |
ENST00000420827.2
ENST00000322244.5 |
UBA6
|
ubiquitin-like modifier activating enzyme 6 |
| chr5_+_99871004 | 0.66 |
ENST00000312637.4
|
FAM174A
|
family with sequence similarity 174, member A |
| chr9_-_77643307 | 0.63 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr7_-_6388537 | 0.62 |
ENST00000313324.4
ENST00000530143.1 |
FAM220A
|
family with sequence similarity 220, member A |
| chr1_+_168148169 | 0.60 |
ENST00000367833.2
|
TIPRL
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr7_-_142247606 | 0.59 |
ENST00000390361.3
|
TRBV7-3
|
T cell receptor beta variable 7-3 |
| chr7_+_89975979 | 0.59 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr5_-_1882858 | 0.58 |
ENST00000511126.1
ENST00000231357.2 |
IRX4
|
iroquois homeobox 4 |
| chr4_+_85504075 | 0.58 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr4_+_170541678 | 0.57 |
ENST00000360642.3
ENST00000512813.1 |
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr3_+_170075436 | 0.56 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr1_-_86861936 | 0.56 |
ENST00000394733.2
ENST00000359242.3 ENST00000294678.2 ENST00000479890.1 ENST00000317336.7 ENST00000370567.1 ENST00000394731.1 ENST00000478286.2 ENST00000370566.3 |
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr4_+_39460659 | 0.56 |
ENST00000513731.1
|
LIAS
|
lipoic acid synthetase |
| chr2_-_192015697 | 0.56 |
ENST00000409995.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr18_+_11851383 | 0.56 |
ENST00000526991.2
|
CHMP1B
|
charged multivesicular body protein 1B |
| chr2_-_105953912 | 0.55 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr5_+_133706865 | 0.55 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr4_+_146402346 | 0.54 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr5_+_44809027 | 0.54 |
ENST00000507110.1
|
MRPS30
|
mitochondrial ribosomal protein S30 |
| chr5_+_68513622 | 0.54 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr3_+_185000729 | 0.53 |
ENST00000448876.1
ENST00000446828.1 ENST00000447637.1 ENST00000424227.1 ENST00000454237.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr5_+_138210919 | 0.52 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr14_+_63671105 | 0.52 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chrX_-_13835461 | 0.52 |
ENST00000316715.4
ENST00000356942.5 |
GPM6B
|
glycoprotein M6B |
| chr18_+_76829441 | 0.52 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr8_+_118533049 | 0.52 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr8_+_17104401 | 0.52 |
ENST00000324815.3
ENST00000518038.1 |
VPS37A
|
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
| chr19_-_51014460 | 0.51 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr1_-_70671216 | 0.51 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr2_-_170681324 | 0.50 |
ENST00000409340.1
|
METTL5
|
methyltransferase like 5 |
| chr7_-_86849836 | 0.50 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr10_-_14920599 | 0.50 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr5_+_147691979 | 0.50 |
ENST00000274565.4
|
SPINK7
|
serine peptidase inhibitor, Kazal type 7 (putative) |
| chrX_-_54070388 | 0.50 |
ENST00000415025.1
|
PHF8
|
PHD finger protein 8 |
| chr5_+_32531893 | 0.49 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr4_-_156297949 | 0.48 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr17_-_8059638 | 0.48 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr4_+_113558272 | 0.47 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr6_-_30523865 | 0.47 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr8_+_26149274 | 0.47 |
ENST00000522535.1
|
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr2_+_28974489 | 0.47 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr1_-_145382434 | 0.46 |
ENST00000610154.1
|
RP11-458D21.1
|
RP11-458D21.1 |
| chr6_-_121655593 | 0.46 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr16_-_81040719 | 0.45 |
ENST00000219400.3
|
CMC2
|
C-x(9)-C motif containing 2 |
| chr19_+_55996316 | 0.45 |
ENST00000205194.4
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr14_-_102976091 | 0.45 |
ENST00000286918.4
|
ANKRD9
|
ankyrin repeat domain 9 |
| chr5_+_158690089 | 0.45 |
ENST00000296786.6
|
UBLCP1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr21_-_30365136 | 0.44 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr6_+_64282447 | 0.44 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr11_-_28129656 | 0.44 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr6_-_26285737 | 0.44 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr13_+_53226963 | 0.44 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr10_-_105156198 | 0.43 |
ENST00000369815.1
ENST00000309579.3 ENST00000337003.4 |
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr7_+_30174668 | 0.43 |
ENST00000415604.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chrX_-_54069253 | 0.43 |
ENST00000425862.1
ENST00000433120.1 |
PHF8
|
PHD finger protein 8 |
| chr14_-_36789783 | 0.42 |
ENST00000605579.1
ENST00000604336.1 ENST00000359527.7 ENST00000603139.1 ENST00000318473.7 |
MBIP
|
MAP3K12 binding inhibitory protein 1 |
| chr2_-_114514181 | 0.42 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr4_-_156298087 | 0.42 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr17_-_8113542 | 0.42 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr22_+_31518938 | 0.42 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr6_+_64281906 | 0.41 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr11_-_73472096 | 0.41 |
ENST00000541588.1
ENST00000336083.3 ENST00000540771.1 ENST00000310653.6 |
RAB6A
|
RAB6A, member RAS oncogene family |
| chr15_-_60771128 | 0.41 |
ENST00000558512.1
ENST00000561114.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr1_-_115124257 | 0.41 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr8_+_38243721 | 0.41 |
ENST00000527334.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr11_+_114271251 | 0.40 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr5_+_112849373 | 0.40 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr11_+_61583968 | 0.40 |
ENST00000517839.1
|
FADS2
|
fatty acid desaturase 2 |
| chr11_-_18343725 | 0.40 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr2_+_190306159 | 0.39 |
ENST00000314761.4
|
WDR75
|
WD repeat domain 75 |
| chr5_+_162887556 | 0.39 |
ENST00000393915.4
ENST00000432118.2 ENST00000358715.3 |
HMMR
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr19_+_1261106 | 0.39 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr16_+_11343475 | 0.39 |
ENST00000572173.1
|
RMI2
|
RecQ mediated genome instability 2 |
| chr1_+_28099700 | 0.39 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr19_+_55996543 | 0.38 |
ENST00000591590.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr6_-_70506963 | 0.38 |
ENST00000370577.3
|
LMBRD1
|
LMBR1 domain containing 1 |
| chr22_+_25003568 | 0.38 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr8_-_82754427 | 0.38 |
ENST00000353788.4
ENST00000520618.1 ENST00000518183.1 ENST00000396330.2 ENST00000519119.1 ENST00000345957.4 |
SNX16
|
sorting nexin 16 |
| chr8_+_117778736 | 0.38 |
ENST00000309822.2
ENST00000357148.3 ENST00000517814.1 ENST00000517820.1 |
UTP23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr6_+_30749649 | 0.38 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr17_+_4046964 | 0.38 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr4_-_77997126 | 0.37 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr10_+_27444268 | 0.37 |
ENST00000375940.4
ENST00000342386.6 |
MASTL
|
microtubule associated serine/threonine kinase-like |
| chr1_+_27719148 | 0.37 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr16_-_3285049 | 0.37 |
ENST00000575948.1
|
ZNF200
|
zinc finger protein 200 |
| chr2_+_24150180 | 0.36 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr2_-_88125286 | 0.36 |
ENST00000327544.6
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr3_+_51851612 | 0.36 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr7_-_86849883 | 0.36 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr20_+_43343886 | 0.36 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr20_-_17949100 | 0.35 |
ENST00000431277.1
|
SNX5
|
sorting nexin 5 |
| chr13_-_60737898 | 0.35 |
ENST00000377908.2
ENST00000400319.1 ENST00000400320.1 ENST00000267215.4 |
DIAPH3
|
diaphanous-related formin 3 |
| chr14_+_23352374 | 0.35 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr19_-_49622348 | 0.35 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr4_-_156298028 | 0.35 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr5_+_61708582 | 0.34 |
ENST00000325324.6
|
IPO11
|
importin 11 |
| chr3_+_16306691 | 0.34 |
ENST00000285083.5
ENST00000605932.1 ENST00000435829.2 |
OXNAD1
|
oxidoreductase NAD-binding domain containing 1 |
| chr3_+_19988736 | 0.34 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr5_-_95297534 | 0.34 |
ENST00000513343.1
ENST00000431061.2 |
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr2_-_55237484 | 0.33 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr11_-_114271139 | 0.33 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr14_+_35515582 | 0.33 |
ENST00000382406.3
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr5_-_133512654 | 0.33 |
ENST00000522552.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr19_+_1954632 | 0.33 |
ENST00000589350.1
|
CSNK1G2
|
casein kinase 1, gamma 2 |
| chr14_+_60715928 | 0.33 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr4_+_44680429 | 0.33 |
ENST00000281543.5
|
GUF1
|
GUF1 GTPase homolog (S. cerevisiae) |
| chr2_+_219745020 | 0.33 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr11_-_63439013 | 0.33 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr18_+_2571510 | 0.33 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr3_+_184056614 | 0.32 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr17_-_63822563 | 0.32 |
ENST00000317442.8
|
CEP112
|
centrosomal protein 112kDa |
| chr8_-_103876383 | 0.32 |
ENST00000347770.4
|
AZIN1
|
antizyme inhibitor 1 |
| chr18_+_76829385 | 0.32 |
ENST00000426216.2
ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B
|
ATPase, class II, type 9B |
| chr20_+_47662805 | 0.32 |
ENST00000262982.2
ENST00000542325.1 |
CSE1L
|
CSE1 chromosome segregation 1-like (yeast) |
| chr1_-_119683251 | 0.32 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_+_82752647 | 0.32 |
ENST00000550058.1
|
METTL25
|
methyltransferase like 25 |
| chr2_+_87140935 | 0.32 |
ENST00000398193.3
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr10_+_103911926 | 0.32 |
ENST00000605788.1
ENST00000405356.1 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr18_-_268019 | 0.31 |
ENST00000261600.6
|
THOC1
|
THO complex 1 |
| chr9_+_132044730 | 0.31 |
ENST00000455981.1
|
RP11-344B5.2
|
RP11-344B5.2 |
| chr14_+_23025534 | 0.31 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr19_+_39786962 | 0.31 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr3_-_154042205 | 0.31 |
ENST00000329463.5
|
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr5_-_118324200 | 0.31 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr4_-_76598544 | 0.31 |
ENST00000515457.1
ENST00000357854.3 |
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_+_7222157 | 0.31 |
ENST00000419721.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr11_+_114271314 | 0.31 |
ENST00000541475.1
|
RBM7
|
RNA binding motif protein 7 |
| chr2_+_183982255 | 0.31 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr14_+_61447832 | 0.30 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr8_+_26149007 | 0.30 |
ENST00000380737.3
ENST00000524169.1 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr16_+_81040794 | 0.30 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr11_+_6502675 | 0.30 |
ENST00000254616.6
ENST00000530751.1 |
TIMM10B
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr11_+_102217936 | 0.30 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr4_+_146019421 | 0.30 |
ENST00000502586.1
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr12_+_82752283 | 0.30 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr5_+_118668846 | 0.30 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr12_-_49412198 | 0.30 |
ENST00000552463.1
|
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr1_-_145382362 | 0.30 |
ENST00000419817.1
ENST00000421937.3 ENST00000433081.2 |
RP11-458D21.1
|
RP11-458D21.1 |
| chr6_-_121655552 | 0.30 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr20_-_17949143 | 0.29 |
ENST00000419004.1
|
SNX5
|
sorting nexin 5 |
| chrX_+_155110956 | 0.29 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr4_-_104119528 | 0.29 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr7_+_23221438 | 0.29 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr18_-_2571210 | 0.29 |
ENST00000577166.1
|
METTL4
|
methyltransferase like 4 |
| chr14_+_93651358 | 0.29 |
ENST00000415050.2
|
TMEM251
|
transmembrane protein 251 |
| chr17_-_6915646 | 0.29 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr11_+_95523823 | 0.29 |
ENST00000538658.1
|
CEP57
|
centrosomal protein 57kDa |
| chr2_+_46769798 | 0.29 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr6_-_136571400 | 0.29 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr19_+_54704610 | 0.28 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr7_+_7222233 | 0.28 |
ENST00000436587.2
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr16_+_70380825 | 0.28 |
ENST00000417604.2
|
DDX19A
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chr1_-_76076793 | 0.28 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr18_-_2571437 | 0.28 |
ENST00000574676.1
ENST00000574538.1 ENST00000319888.6 |
METTL4
|
methyltransferase like 4 |
| chr17_+_25621102 | 0.28 |
ENST00000581440.1
ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1
|
WD repeat and SOCS box containing 1 |
| chr16_-_3285144 | 0.28 |
ENST00000431561.3
ENST00000396870.4 |
ZNF200
|
zinc finger protein 200 |
| chr1_+_242011468 | 0.28 |
ENST00000366548.3
|
EXO1
|
exonuclease 1 |
| chr2_+_108443388 | 0.28 |
ENST00000354986.4
ENST00000408999.3 |
RGPD4
|
RANBP2-like and GRIP domain containing 4 |
| chr14_+_38065052 | 0.28 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr13_+_34392200 | 0.27 |
ENST00000434425.1
|
RFC3
|
replication factor C (activator 1) 3, 38kDa |
| chr3_+_44379944 | 0.27 |
ENST00000396078.3
ENST00000342649.4 |
TCAIM
|
T cell activation inhibitor, mitochondrial |
| chr1_-_150533969 | 0.27 |
ENST00000369035.2
|
ADAMTSL4-AS1
|
ADAMTSL4 antisense RNA 1 |
| chr19_-_9695169 | 0.27 |
ENST00000586602.1
|
ZNF121
|
zinc finger protein 121 |
| chr8_-_17104356 | 0.27 |
ENST00000361272.4
ENST00000523917.1 |
CNOT7
|
CCR4-NOT transcription complex, subunit 7 |
| chr17_-_15902903 | 0.27 |
ENST00000486655.1
|
ZSWIM7
|
zinc finger, SWIM-type containing 7 |
| chr19_-_50979981 | 0.27 |
ENST00000595790.1
ENST00000600100.1 |
FAM71E1
|
family with sequence similarity 71, member E1 |
| chr5_+_41904431 | 0.27 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr4_-_73935409 | 0.27 |
ENST00000507544.2
ENST00000295890.4 |
COX18
|
COX18 cytochrome C oxidase assembly factor |
| chr3_-_12587055 | 0.27 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr3_+_99979828 | 0.27 |
ENST00000485687.1
ENST00000344949.5 ENST00000394144.4 |
TBC1D23
|
TBC1 domain family, member 23 |
| chr18_-_47013586 | 0.27 |
ENST00000318240.3
ENST00000579820.1 |
C18orf32
|
chromosome 18 open reading frame 32 |
| chr12_+_108079509 | 0.27 |
ENST00000412830.3
ENST00000547995.1 |
PWP1
|
PWP1 homolog (S. cerevisiae) |
| chr4_-_79860506 | 0.27 |
ENST00000295462.3
ENST00000380645.4 ENST00000512733.1 |
PAQR3
|
progestin and adipoQ receptor family member III |
| chr17_-_77967433 | 0.26 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr12_+_97300995 | 0.26 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr11_-_18343669 | 0.26 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chrX_-_77150985 | 0.26 |
ENST00000358075.6
|
MAGT1
|
magnesium transporter 1 |
| chr17_+_42385927 | 0.26 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr20_+_55099542 | 0.26 |
ENST00000371328.3
|
FAM209A
|
family with sequence similarity 209, member A |
| chr14_+_60716159 | 0.26 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF2_ATF1_ATF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.2 | 1.0 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.2 | 0.9 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.2 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.2 | 0.6 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.2 | 2.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.7 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 0.6 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.2 | 0.9 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.4 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.4 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.5 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.3 | GO:0070510 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.5 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 0.3 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.1 | 0.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.9 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 1.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.7 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 1.0 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.3 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.1 | 0.7 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.5 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.2 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.6 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.2 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.1 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.3 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 0.8 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.3 | GO:0045829 | negative regulation of isotype switching(GO:0045829) |
| 0.1 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.4 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.8 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 1.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.1 | 0.1 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.2 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.1 | 0.6 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.1 | GO:1903659 | complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.1 | 0.6 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.6 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 0.1 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.2 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 0.4 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.6 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 0.4 | GO:1900003 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.5 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.2 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 2.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.2 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.0 | 0.2 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.0 | 0.0 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.4 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.2 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.0 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.4 | GO:0007135 | meiosis II(GO:0007135) |
| 0.0 | 0.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0046833 | snRNA export from nucleus(GO:0006408) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.7 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.4 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 1.1 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.2 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0050992 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
| 0.0 | 0.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0045345 | positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0043103 | hypoxanthine salvage(GO:0043103) |
| 0.0 | 0.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.3 | GO:0060701 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 1.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.0 | GO:0050658 | nucleic acid transport(GO:0050657) RNA transport(GO:0050658) establishment of RNA localization(GO:0051236) |
| 0.0 | 0.9 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0030099 | myeloid cell differentiation(GO:0030099) |
| 0.0 | 0.3 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.5 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.0 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.3 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.8 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.2 | GO:0030047 | actin modification(GO:0030047) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.0 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) striated muscle adaptation(GO:0014888) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.7 | GO:0000732 | strand displacement(GO:0000732) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:0034130 | toll-like receptor 1 signaling pathway(GO:0034130) |
| 0.0 | 0.2 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.6 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.0 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 0.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.0 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.9 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.0 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0051106 | positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:0042533 | tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.0 | 0.1 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.0 | GO:0051384 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) |
| 0.0 | 0.4 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:1902218 | regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.0 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.0 | 0.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.0 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.0 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0009407 | toxin catabolic process(GO:0009407) secondary metabolite catabolic process(GO:0090487) |
| 0.0 | 0.0 | GO:0036337 | Fas signaling pathway(GO:0036337) regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.2 | 0.6 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.2 | 0.7 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.2 | 1.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.3 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.1 | 2.7 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.3 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 0.3 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 0.7 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.6 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.2 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 0.3 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.9 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.5 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.2 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.2 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.4 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.8 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.0 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.0 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005642 | annulate lamellae(GO:0005642) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.2 | 0.9 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.2 | 0.7 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.2 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.7 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.5 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 2.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.3 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.1 | 0.5 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0016213 | linoleoyl-CoA desaturase activity(GO:0016213) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.2 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.2 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.7 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.2 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.9 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.3 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.8 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.7 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.4 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 1.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.3 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 1.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.0 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.0 | GO:0045485 | omega-6 fatty acid desaturase activity(GO:0045485) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.1 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.0 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 1.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.0 | GO:0034594 | phosphatidylinositol trisphosphate phosphatase activity(GO:0034594) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.4 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.1 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |