Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
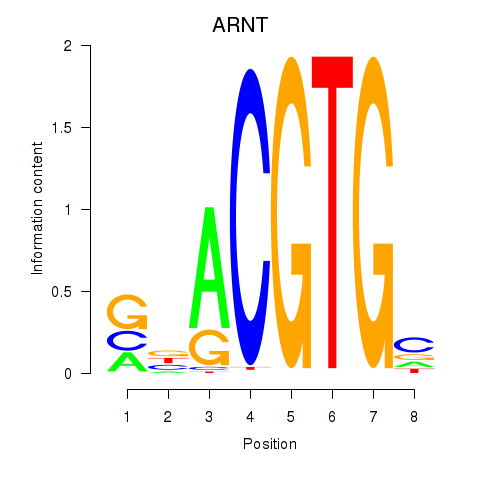
Results for ARNT
Z-value: 0.29
Transcription factors associated with ARNT
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARNT
|
ENSG00000143437.16 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARNT | hg19_v2_chr1_-_150849174_150849200 | 0.47 | 3.4e-01 | Click! |
Activity profile of ARNT motif
Sorted Z-values of ARNT motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_100435535 | 0.16 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr1_+_100436065 | 0.14 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr3_+_19988736 | 0.13 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr15_+_45722727 | 0.12 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr10_-_126849626 | 0.12 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr8_+_95907993 | 0.11 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr6_+_89791507 | 0.10 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr8_+_109455830 | 0.10 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr1_-_94344686 | 0.10 |
ENST00000528680.1
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr12_+_64798095 | 0.10 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr3_-_145878954 | 0.10 |
ENST00000282903.5
ENST00000360060.3 |
PLOD2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr2_-_220117867 | 0.10 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr1_+_27669719 | 0.09 |
ENST00000473280.1
|
SYTL1
|
synaptotagmin-like 1 |
| chr1_+_92495528 | 0.08 |
ENST00000370383.4
|
EPHX4
|
epoxide hydrolase 4 |
| chr4_-_4291761 | 0.08 |
ENST00000513174.1
|
LYAR
|
Ly1 antibody reactive |
| chr20_-_3219359 | 0.08 |
ENST00000437836.2
|
SLC4A11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr12_-_120907374 | 0.08 |
ENST00000550458.1
|
SRSF9
|
serine/arginine-rich splicing factor 9 |
| chr12_-_51422017 | 0.08 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr7_+_23637118 | 0.08 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr10_+_122216707 | 0.08 |
ENST00000427079.1
ENST00000541332.1 |
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A |
| chr22_-_24181174 | 0.08 |
ENST00000318109.7
ENST00000406855.3 ENST00000404056.1 ENST00000476077.1 |
DERL3
|
derlin 3 |
| chr2_-_10587897 | 0.08 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr8_+_26240666 | 0.08 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_-_64010006 | 0.08 |
ENST00000555899.1
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr7_-_139876734 | 0.07 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr20_+_56964253 | 0.07 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr18_-_33077868 | 0.07 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr3_-_195808952 | 0.07 |
ENST00000540528.1
ENST00000392396.3 ENST00000535031.1 ENST00000420415.1 |
TFRC
|
transferrin receptor |
| chr3_+_66271410 | 0.07 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr19_+_10765614 | 0.07 |
ENST00000589283.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr3_-_195808980 | 0.07 |
ENST00000360110.4
|
TFRC
|
transferrin receptor |
| chr19_-_663147 | 0.07 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr5_-_171881362 | 0.07 |
ENST00000519643.1
|
SH3PXD2B
|
SH3 and PX domains 2B |
| chr12_-_31478428 | 0.07 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr16_-_81129845 | 0.07 |
ENST00000569885.1
ENST00000566566.1 |
GCSH
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr9_-_135545380 | 0.07 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chrX_+_77359726 | 0.07 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr2_-_165477971 | 0.06 |
ENST00000446413.2
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr22_-_39548443 | 0.06 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr17_+_27052892 | 0.06 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr6_+_73331918 | 0.06 |
ENST00000402622.2
ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr7_-_130597935 | 0.06 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr2_-_232328867 | 0.06 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr7_-_127032363 | 0.06 |
ENST00000393312.1
|
ZNF800
|
zinc finger protein 800 |
| chr7_-_127032114 | 0.06 |
ENST00000436992.1
|
ZNF800
|
zinc finger protein 800 |
| chr9_+_133569108 | 0.06 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr17_+_41132564 | 0.06 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr7_-_105925367 | 0.06 |
ENST00000354289.4
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr3_-_156272924 | 0.06 |
ENST00000467789.1
ENST00000265044.2 |
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr17_+_40440481 | 0.06 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr19_+_10765699 | 0.06 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr15_-_30261066 | 0.06 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr7_+_26331541 | 0.06 |
ENST00000416246.1
ENST00000338523.4 ENST00000412416.1 |
SNX10
|
sorting nexin 10 |
| chr2_+_171627597 | 0.06 |
ENST00000429172.1
ENST00000426475.1 |
AC007405.6
|
AC007405.6 |
| chrX_+_51075658 | 0.06 |
ENST00000356450.2
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr1_+_47799446 | 0.06 |
ENST00000371873.5
|
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr6_-_7313381 | 0.06 |
ENST00000489567.1
ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1
|
signal sequence receptor, alpha |
| chr1_-_246729544 | 0.06 |
ENST00000544618.1
ENST00000366514.4 |
TFB2M
|
transcription factor B2, mitochondrial |
| chr18_-_33077556 | 0.06 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr7_-_19157248 | 0.06 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr2_+_28113583 | 0.06 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr7_-_105925558 | 0.06 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr12_-_49523896 | 0.06 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr1_-_6269448 | 0.06 |
ENST00000465335.1
|
RPL22
|
ribosomal protein L22 |
| chr1_+_27719148 | 0.06 |
ENST00000374024.3
|
GPR3
|
G protein-coupled receptor 3 |
| chr16_+_66914264 | 0.06 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr16_-_71323271 | 0.06 |
ENST00000565850.1
ENST00000568910.1 ENST00000434935.2 ENST00000338099.5 |
CMTR2
|
cap methyltransferase 2 |
| chr7_-_139876812 | 0.05 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr2_-_136743039 | 0.05 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr10_+_112257596 | 0.05 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr1_-_6259641 | 0.05 |
ENST00000234875.4
|
RPL22
|
ribosomal protein L22 |
| chr5_-_172198190 | 0.05 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chr1_+_209848749 | 0.05 |
ENST00000367029.4
|
G0S2
|
G0/G1switch 2 |
| chr2_-_148778323 | 0.05 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr20_-_17949100 | 0.05 |
ENST00000431277.1
|
SNX5
|
sorting nexin 5 |
| chr4_+_186317133 | 0.05 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr3_-_160823158 | 0.05 |
ENST00000392779.2
ENST00000392780.1 ENST00000494173.1 |
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr12_+_6977323 | 0.05 |
ENST00000462761.1
|
TPI1
|
triosephosphate isomerase 1 |
| chr1_+_65613852 | 0.05 |
ENST00000327299.7
|
AK4
|
adenylate kinase 4 |
| chr6_+_43739697 | 0.05 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chrX_+_77359671 | 0.05 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr4_-_103266626 | 0.05 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_159825137 | 0.05 |
ENST00000368102.1
|
C1orf204
|
chromosome 1 open reading frame 204 |
| chr19_+_1261106 | 0.05 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr17_-_27277615 | 0.05 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr1_-_26232522 | 0.05 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr8_+_104426942 | 0.05 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr1_-_26232951 | 0.05 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr1_-_6259613 | 0.05 |
ENST00000465387.1
|
RPL22
|
ribosomal protein L22 |
| chr3_-_156272872 | 0.05 |
ENST00000476217.1
|
SSR3
|
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr2_-_157189180 | 0.05 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr15_-_65281775 | 0.05 |
ENST00000433215.2
ENST00000558415.1 ENST00000557795.1 |
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr6_-_83775489 | 0.05 |
ENST00000369747.3
|
UBE3D
|
ubiquitin protein ligase E3D |
| chr13_-_52027134 | 0.05 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr2_+_216176761 | 0.05 |
ENST00000540518.1
ENST00000435675.1 |
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr15_-_65282274 | 0.05 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chrX_-_41782683 | 0.05 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr10_-_22292675 | 0.05 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr1_+_44445549 | 0.05 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr5_-_38845812 | 0.05 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr3_+_100211412 | 0.04 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr3_-_56717246 | 0.04 |
ENST00000355628.5
|
FAM208A
|
family with sequence similarity 208, member A |
| chr5_-_114961858 | 0.04 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr22_+_31742875 | 0.04 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr5_+_49962772 | 0.04 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_26240414 | 0.04 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr1_+_235490659 | 0.04 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr17_+_49337881 | 0.04 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr7_-_138720763 | 0.04 |
ENST00000275766.1
|
ZC3HAV1L
|
zinc finger CCCH-type, antiviral 1-like |
| chr2_+_30370382 | 0.04 |
ENST00000402708.1
|
YPEL5
|
yippee-like 5 (Drosophila) |
| chr6_+_33257346 | 0.04 |
ENST00000374606.5
ENST00000374610.2 ENST00000374607.1 |
PFDN6
|
prefoldin subunit 6 |
| chr21_-_34915084 | 0.04 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr3_+_25831567 | 0.04 |
ENST00000280701.3
ENST00000420173.2 |
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr14_+_74004051 | 0.04 |
ENST00000557556.1
|
ACOT1
|
acyl-CoA thioesterase 1 |
| chr11_+_18416133 | 0.04 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr2_-_220118631 | 0.04 |
ENST00000248437.4
|
TUBA4A
|
tubulin, alpha 4a |
| chr10_+_98592009 | 0.04 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr7_+_30185496 | 0.04 |
ENST00000455738.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr9_+_74526384 | 0.04 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr20_+_35201993 | 0.04 |
ENST00000373872.4
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr3_+_122785895 | 0.04 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chrX_-_119693745 | 0.04 |
ENST00000371323.2
|
CUL4B
|
cullin 4B |
| chr20_-_17949143 | 0.04 |
ENST00000419004.1
|
SNX5
|
sorting nexin 5 |
| chr1_+_47799542 | 0.04 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr15_-_65282232 | 0.04 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_-_6269304 | 0.04 |
ENST00000471204.1
|
RPL22
|
ribosomal protein L22 |
| chr3_+_133292574 | 0.04 |
ENST00000264993.3
|
CDV3
|
CDV3 homolog (mouse) |
| chr8_-_37756972 | 0.04 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr9_+_112542591 | 0.04 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr2_+_32288657 | 0.04 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr3_+_133292759 | 0.04 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr11_-_64014379 | 0.04 |
ENST00000309318.3
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr5_+_49962495 | 0.04 |
ENST00000515175.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr15_-_60884706 | 0.04 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr1_-_26233423 | 0.04 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr20_+_35201857 | 0.04 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr9_+_112542572 | 0.04 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr14_+_64854958 | 0.04 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr11_-_123525289 | 0.04 |
ENST00000392770.2
ENST00000299333.3 ENST00000530277.1 |
SCN3B
|
sodium channel, voltage-gated, type III, beta subunit |
| chr1_+_214454492 | 0.04 |
ENST00000366957.5
ENST00000415093.2 |
SMYD2
|
SET and MYND domain containing 2 |
| chr17_-_27053216 | 0.04 |
ENST00000292090.3
|
TLCD1
|
TLC domain containing 1 |
| chr11_-_122931881 | 0.04 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr17_-_48207115 | 0.04 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chr2_+_32288725 | 0.04 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr14_+_65453432 | 0.04 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr5_+_154238149 | 0.04 |
ENST00000519430.1
ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8
|
CCR4-NOT transcription complex, subunit 8 |
| chr7_-_6523688 | 0.04 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr5_+_34656569 | 0.04 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr11_+_18416103 | 0.04 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr20_-_6103666 | 0.04 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr5_-_36242119 | 0.03 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr8_-_90996459 | 0.03 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chr7_+_23636992 | 0.03 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr2_-_148778258 | 0.03 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr7_-_108166505 | 0.03 |
ENST00000426128.2
ENST00000427008.1 ENST00000388728.5 ENST00000257694.8 ENST00000422087.1 ENST00000453144.1 ENST00000436062.1 |
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr1_+_228353495 | 0.03 |
ENST00000366711.3
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr19_-_47353547 | 0.03 |
ENST00000601498.1
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr3_+_112280857 | 0.03 |
ENST00000492406.1
ENST00000468642.1 |
SLC35A5
|
solute carrier family 35, member A5 |
| chr2_+_216176540 | 0.03 |
ENST00000236959.9
|
ATIC
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr17_-_40075219 | 0.03 |
ENST00000537919.1
ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY
|
ATP citrate lyase |
| chr14_-_53162361 | 0.03 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr18_+_9913977 | 0.03 |
ENST00000400000.2
ENST00000340541.4 |
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
| chr19_-_47354023 | 0.03 |
ENST00000601649.1
ENST00000599990.1 ENST00000352203.4 |
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr7_-_17980091 | 0.03 |
ENST00000409389.1
ENST00000409604.1 ENST00000428135.3 |
SNX13
|
sorting nexin 13 |
| chr20_+_30946106 | 0.03 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr2_+_64069459 | 0.03 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr18_-_55288973 | 0.03 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr7_-_128045984 | 0.03 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr1_-_111991850 | 0.03 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr20_+_2633269 | 0.03 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr10_-_103543145 | 0.03 |
ENST00000370110.5
|
NPM3
|
nucleophosmin/nucleoplasmin 3 |
| chr2_-_73460334 | 0.03 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr5_+_80256453 | 0.03 |
ENST00000265080.4
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2 |
| chr17_+_40985407 | 0.03 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr12_+_110906169 | 0.03 |
ENST00000377673.5
|
FAM216A
|
family with sequence similarity 216, member A |
| chr7_+_56119323 | 0.03 |
ENST00000275603.4
ENST00000335503.3 ENST00000540286.1 |
CCT6A
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr1_-_159894319 | 0.03 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr6_+_151187615 | 0.03 |
ENST00000441122.1
ENST00000423867.1 |
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_9781068 | 0.03 |
ENST00000500698.1
|
RP11-540A21.2
|
RP11-540A21.2 |
| chr6_+_64281906 | 0.03 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr17_-_42907564 | 0.03 |
ENST00000592524.1
|
GJC1
|
gap junction protein, gamma 1, 45kDa |
| chr8_-_90996837 | 0.03 |
ENST00000519426.1
ENST00000265433.3 |
NBN
|
nibrin |
| chr2_+_75061108 | 0.03 |
ENST00000290573.2
|
HK2
|
hexokinase 2 |
| chr20_+_56964169 | 0.03 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_+_10353780 | 0.03 |
ENST00000449913.2
ENST00000503788.1 ENST00000274140.5 |
MARCH6
|
membrane-associated ring finger (C3HC4) 6, E3 ubiquitin protein ligase |
| chr18_-_33077942 | 0.03 |
ENST00000334598.7
|
INO80C
|
INO80 complex subunit C |
| chr14_-_69263043 | 0.03 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr2_+_64069240 | 0.03 |
ENST00000497883.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chrX_+_16804544 | 0.03 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr9_-_123639304 | 0.03 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr5_+_98264867 | 0.03 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr4_+_85504075 | 0.03 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr20_+_31407692 | 0.03 |
ENST00000375571.5
|
MAPRE1
|
microtubule-associated protein, RP/EB family, member 1 |
| chr6_+_84569359 | 0.03 |
ENST00000369681.5
ENST00000369679.4 |
CYB5R4
|
cytochrome b5 reductase 4 |
| chrX_-_13956497 | 0.03 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr15_+_44084503 | 0.03 |
ENST00000409960.2
ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2
|
small EDRK-rich factor 2 |
| chr7_+_73082152 | 0.03 |
ENST00000324941.4
ENST00000451519.1 |
VPS37D
|
vacuolar protein sorting 37 homolog D (S. cerevisiae) |
| chr16_+_2867228 | 0.03 |
ENST00000005995.3
ENST00000574813.1 |
PRSS21
|
protease, serine, 21 (testisin) |
| chr17_-_67323385 | 0.03 |
ENST00000588665.1
|
ABCA5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr3_-_57583185 | 0.03 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARNT
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.0 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.0 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:1901910 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.0 | GO:0009346 | citrate lyase complex(GO:0009346) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0004641 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.0 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0052843 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |