Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
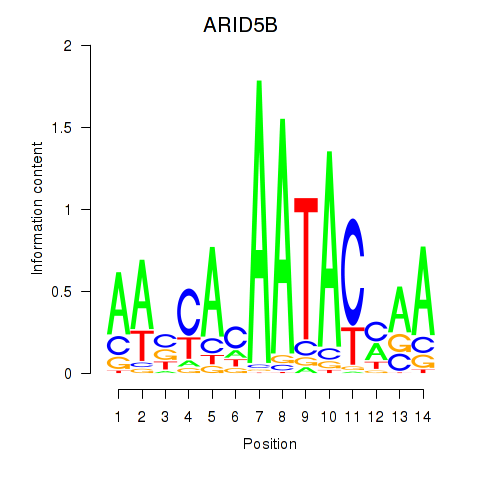
Results for ARID5B
Z-value: 0.67
Transcription factors associated with ARID5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5B
|
ENSG00000150347.10 | AT-rich interaction domain 5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5B | hg19_v2_chr10_+_63808970_63808990 | 0.85 | 3.3e-02 | Click! |
Activity profile of ARID5B motif
Sorted Z-values of ARID5B motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_74178582 | 0.34 |
ENST00000531854.1
ENST00000526855.1 ENST00000529425.1 ENST00000310128.4 ENST00000532569.1 |
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3 |
| chr12_-_15038779 | 0.29 |
ENST00000228938.5
ENST00000539261.1 |
MGP
|
matrix Gla protein |
| chr1_+_95975672 | 0.28 |
ENST00000440116.2
ENST00000456933.1 |
RP11-286B14.1
|
RP11-286B14.1 |
| chr8_-_72268721 | 0.27 |
ENST00000419131.1
ENST00000388743.2 |
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr18_-_25616519 | 0.26 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr9_-_116065551 | 0.23 |
ENST00000297894.5
|
RNF183
|
ring finger protein 183 |
| chr11_-_102714534 | 0.22 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr9_+_42671887 | 0.22 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr5_+_32788945 | 0.21 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chr1_-_161337662 | 0.21 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr15_+_36994210 | 0.20 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr12_-_124456598 | 0.20 |
ENST00000539761.1
ENST00000539551.1 |
CCDC92
|
coiled-coil domain containing 92 |
| chr12_-_118796910 | 0.19 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr7_+_116654958 | 0.17 |
ENST00000449366.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr2_+_223725723 | 0.17 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr1_-_205391178 | 0.16 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr18_+_9102669 | 0.16 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr1_-_117021430 | 0.15 |
ENST00000423907.1
ENST00000434879.1 ENST00000443219.1 |
RP4-655J12.4
|
RP4-655J12.4 |
| chr7_+_116660246 | 0.15 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr8_-_72268889 | 0.15 |
ENST00000388742.4
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr8_+_94752349 | 0.15 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr8_-_72268968 | 0.15 |
ENST00000388740.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr12_-_118796971 | 0.15 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr8_-_101321584 | 0.15 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr1_-_85742773 | 0.15 |
ENST00000370580.1
|
BCL10
|
B-cell CLL/lymphoma 10 |
| chr5_-_64920115 | 0.14 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr7_+_36450169 | 0.14 |
ENST00000428612.1
|
ANLN
|
anillin, actin binding protein |
| chr12_+_51236703 | 0.13 |
ENST00000551456.1
ENST00000398458.3 |
TMPRSS12
|
transmembrane (C-terminal) protease, serine 12 |
| chrX_+_86772787 | 0.13 |
ENST00000373114.4
|
KLHL4
|
kelch-like family member 4 |
| chr7_-_56119156 | 0.13 |
ENST00000421312.1
ENST00000416592.1 |
PSPH
|
phosphoserine phosphatase |
| chr9_+_71736177 | 0.12 |
ENST00000606364.1
ENST00000453658.2 |
TJP2
|
tight junction protein 2 |
| chr4_+_147096837 | 0.12 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr16_-_3422283 | 0.12 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr8_-_116504448 | 0.12 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr8_+_92114873 | 0.12 |
ENST00000343709.3
ENST00000448384.2 |
LRRC69
|
leucine rich repeat containing 69 |
| chr6_+_122720681 | 0.12 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr10_-_75351088 | 0.11 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr12_+_79258444 | 0.11 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr11_-_73882057 | 0.11 |
ENST00000334126.7
ENST00000313663.7 |
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr13_+_53216565 | 0.11 |
ENST00000357495.2
|
HNRNPA1L2
|
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr19_+_38307999 | 0.11 |
ENST00000589653.1
ENST00000590433.1 |
CTD-2554C21.2
|
CTD-2554C21.2 |
| chr3_+_32726774 | 0.10 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr10_+_4828815 | 0.10 |
ENST00000533295.1
|
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr1_+_78245466 | 0.10 |
ENST00000477627.2
|
FAM73A
|
family with sequence similarity 73, member A |
| chr6_+_126661253 | 0.10 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr20_-_9819674 | 0.10 |
ENST00000378429.3
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr17_-_76732928 | 0.10 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr11_-_73882249 | 0.10 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr22_+_19467261 | 0.10 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr1_-_114414316 | 0.10 |
ENST00000528414.1
ENST00000538253.1 ENST00000460620.1 ENST00000420377.2 ENST00000525799.1 ENST00000359785.5 |
PTPN22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr1_+_186798073 | 0.10 |
ENST00000367466.3
ENST00000442353.2 |
PLA2G4A
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr12_+_128399965 | 0.09 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr12_-_8815299 | 0.09 |
ENST00000535336.1
|
MFAP5
|
microfibrillar associated protein 5 |
| chr3_-_98241358 | 0.09 |
ENST00000503004.1
ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1
|
claudin domain containing 1 |
| chr12_-_8815215 | 0.09 |
ENST00000544889.1
ENST00000543369.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chrX_-_101410762 | 0.09 |
ENST00000543160.1
ENST00000333643.3 |
BEX5
|
brain expressed, X-linked 5 |
| chr1_+_104159999 | 0.09 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr1_-_144994840 | 0.09 |
ENST00000369351.3
ENST00000369349.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr2_+_27760247 | 0.09 |
ENST00000447166.1
|
AC109829.1
|
Uncharacterized protein |
| chr22_+_42086547 | 0.09 |
ENST00000402966.1
|
C22orf46
|
chromosome 22 open reading frame 46 |
| chr4_+_71588372 | 0.09 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_+_32608566 | 0.08 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr5_+_173763250 | 0.08 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr14_+_85994943 | 0.08 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr3_+_32726620 | 0.08 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr6_-_30080876 | 0.08 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr11_+_1093318 | 0.08 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr3_-_139396853 | 0.08 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr12_+_128399917 | 0.08 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr7_+_143080063 | 0.08 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr1_-_224624730 | 0.08 |
ENST00000445239.1
|
WDR26
|
WD repeat domain 26 |
| chr9_+_35806082 | 0.08 |
ENST00000447210.1
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr12_+_56325231 | 0.08 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr6_-_30080863 | 0.08 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr22_-_29107919 | 0.08 |
ENST00000434810.1
ENST00000456369.1 |
CHEK2
|
checkpoint kinase 2 |
| chr7_+_149535455 | 0.07 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr1_-_144994909 | 0.07 |
ENST00000369347.4
ENST00000369354.3 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr12_-_8814669 | 0.07 |
ENST00000535411.1
ENST00000540087.1 |
MFAP5
|
microfibrillar associated protein 5 |
| chr12_+_79258547 | 0.07 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr6_+_80341000 | 0.07 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr8_-_11710979 | 0.07 |
ENST00000415599.2
|
CTSB
|
cathepsin B |
| chr20_-_45980621 | 0.07 |
ENST00000446894.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr18_-_53303123 | 0.07 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr17_-_21454898 | 0.07 |
ENST00000391411.5
ENST00000412778.3 |
C17orf51
|
chromosome 17 open reading frame 51 |
| chr11_+_73882311 | 0.07 |
ENST00000398427.4
ENST00000544401.1 |
PPME1
|
protein phosphatase methylesterase 1 |
| chr12_-_6809958 | 0.07 |
ENST00000320591.5
ENST00000534837.1 |
PIANP
|
PILR alpha associated neural protein |
| chr12_-_15815626 | 0.07 |
ENST00000540613.1
|
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_-_144995074 | 0.07 |
ENST00000534536.1
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr5_+_68462837 | 0.07 |
ENST00000256442.5
|
CCNB1
|
cyclin B1 |
| chr1_+_171107241 | 0.07 |
ENST00000236166.3
|
FMO6P
|
flavin containing monooxygenase 6 pseudogene |
| chr19_+_58180303 | 0.06 |
ENST00000318203.5
|
ZSCAN4
|
zinc finger and SCAN domain containing 4 |
| chr10_-_69835001 | 0.06 |
ENST00000513996.1
ENST00000412272.2 ENST00000395198.3 ENST00000492996.2 |
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr14_+_32798547 | 0.06 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr2_+_197577841 | 0.06 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr7_+_116654935 | 0.06 |
ENST00000432298.1
ENST00000422922.1 |
ST7
|
suppression of tumorigenicity 7 |
| chr7_-_117512264 | 0.06 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chr1_-_168698433 | 0.06 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr7_-_56119238 | 0.06 |
ENST00000275605.3
ENST00000395471.3 |
PSPH
|
phosphoserine phosphatase |
| chr9_+_114287433 | 0.06 |
ENST00000358151.4
ENST00000355824.3 ENST00000374374.3 ENST00000309235.5 |
ZNF483
|
zinc finger protein 483 |
| chr22_-_22090043 | 0.05 |
ENST00000403503.1
|
YPEL1
|
yippee-like 1 (Drosophila) |
| chr10_+_126150369 | 0.05 |
ENST00000392757.4
ENST00000368842.5 ENST00000368839.1 |
LHPP
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr3_-_98241598 | 0.05 |
ENST00000513287.1
ENST00000514537.1 ENST00000508071.1 ENST00000507944.1 |
CLDND1
|
claudin domain containing 1 |
| chr8_+_55466915 | 0.05 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr1_-_76076793 | 0.05 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr10_-_69835099 | 0.05 |
ENST00000373700.4
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr18_+_20513278 | 0.05 |
ENST00000327155.5
|
RBBP8
|
retinoblastoma binding protein 8 |
| chr2_+_223725652 | 0.05 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr19_-_56826157 | 0.05 |
ENST00000592509.1
ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A
|
zinc finger and SCAN domain containing 5A |
| chr1_+_158975744 | 0.05 |
ENST00000426592.2
|
IFI16
|
interferon, gamma-inducible protein 16 |
| chr5_+_137225125 | 0.04 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr4_-_159094194 | 0.04 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr12_-_10007448 | 0.04 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr1_+_104293028 | 0.04 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr1_+_61330931 | 0.04 |
ENST00000371191.1
|
NFIA
|
nuclear factor I/A |
| chr2_-_114300213 | 0.04 |
ENST00000446595.1
ENST00000416105.1 ENST00000450636.1 ENST00000416758.1 |
RP11-395L14.4
|
RP11-395L14.4 |
| chrX_-_49089771 | 0.04 |
ENST00000376251.1
ENST00000323022.5 ENST00000376265.2 |
CACNA1F
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr8_+_55467072 | 0.04 |
ENST00000602362.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr18_+_21032781 | 0.04 |
ENST00000339486.3
|
RIOK3
|
RIO kinase 3 |
| chr10_-_128210005 | 0.04 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr14_-_69263043 | 0.04 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr12_-_122879969 | 0.04 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr7_+_138145145 | 0.04 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr19_+_52956798 | 0.04 |
ENST00000421239.2
|
ZNF578
|
zinc finger protein 578 |
| chr12_+_8309630 | 0.04 |
ENST00000396570.3
|
ZNF705A
|
zinc finger protein 705A |
| chr3_+_161214596 | 0.04 |
ENST00000327928.4
|
OTOL1
|
otolin 1 |
| chrX_-_11369656 | 0.04 |
ENST00000413512.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr17_-_3599696 | 0.04 |
ENST00000225328.5
|
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr10_-_69834973 | 0.04 |
ENST00000395187.2
|
HERC4
|
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
| chr11_-_33774944 | 0.03 |
ENST00000532057.1
ENST00000531080.1 |
FBXO3
|
F-box protein 3 |
| chr9_-_32552551 | 0.03 |
ENST00000360538.2
ENST00000379858.1 |
TOPORS
|
topoisomerase I binding, arginine/serine-rich, E3 ubiquitin protein ligase |
| chrX_-_33229636 | 0.03 |
ENST00000357033.4
|
DMD
|
dystrophin |
| chr17_-_3599492 | 0.03 |
ENST00000435558.1
ENST00000345901.3 |
P2RX5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_-_15540055 | 0.03 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr5_-_88179017 | 0.03 |
ENST00000514028.1
ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C
|
myocyte enhancer factor 2C |
| chr3_+_196594727 | 0.03 |
ENST00000445299.2
ENST00000323460.5 ENST00000419026.1 |
SENP5
|
SUMO1/sentrin specific peptidase 5 |
| chr4_-_89978299 | 0.03 |
ENST00000511976.1
ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr5_-_179047881 | 0.03 |
ENST00000521173.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr6_+_90192974 | 0.03 |
ENST00000520458.1
|
ANKRD6
|
ankyrin repeat domain 6 |
| chr7_-_24797546 | 0.03 |
ENST00000414428.1
ENST00000419307.1 ENST00000342947.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr3_+_190333097 | 0.03 |
ENST00000412080.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr18_+_72166564 | 0.03 |
ENST00000583216.1
ENST00000581912.1 ENST00000582589.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr5_+_125798031 | 0.03 |
ENST00000502348.1
|
GRAMD3
|
GRAM domain containing 3 |
| chrX_-_11308598 | 0.03 |
ENST00000380717.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr4_-_71532207 | 0.02 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr14_+_96968707 | 0.02 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr1_+_6615241 | 0.02 |
ENST00000333172.6
ENST00000328191.4 ENST00000351136.3 |
TAS1R1
|
taste receptor, type 1, member 1 |
| chr4_+_40201954 | 0.02 |
ENST00000511121.1
|
RHOH
|
ras homolog family member H |
| chr7_-_37488834 | 0.02 |
ENST00000310758.4
|
ELMO1
|
engulfment and cell motility 1 |
| chr1_-_198906528 | 0.02 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr12_+_120933904 | 0.02 |
ENST00000550178.1
ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1
|
dynein, light chain, LC8-type 1 |
| chr14_-_31856397 | 0.02 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr12_+_56390964 | 0.02 |
ENST00000356124.4
ENST00000266971.3 ENST00000394115.2 ENST00000547586.1 ENST00000552258.1 ENST00000548274.1 ENST00000546833.1 |
SUOX
|
sulfite oxidase |
| chr1_-_115292591 | 0.02 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr3_-_74570291 | 0.02 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr1_+_200011711 | 0.02 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr4_+_189060573 | 0.02 |
ENST00000332517.3
|
TRIML1
|
tripartite motif family-like 1 |
| chr2_-_178753465 | 0.02 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr12_+_56324756 | 0.02 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr1_-_76076759 | 0.02 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr3_+_111805182 | 0.02 |
ENST00000430855.1
ENST00000431717.2 ENST00000264848.5 |
C3orf52
|
chromosome 3 open reading frame 52 |
| chr9_+_116267536 | 0.02 |
ENST00000374136.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr6_+_32812568 | 0.02 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr1_-_43855479 | 0.01 |
ENST00000290663.6
ENST00000372457.4 |
MED8
|
mediator complex subunit 8 |
| chr6_+_42896865 | 0.01 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr3_-_79816965 | 0.01 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_+_137225158 | 0.01 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr10_-_99030395 | 0.01 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr1_+_150039369 | 0.01 |
ENST00000369130.3
ENST00000369128.5 |
VPS45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr15_+_51669444 | 0.01 |
ENST00000396399.2
|
GLDN
|
gliomedin |
| chrX_+_54466829 | 0.01 |
ENST00000375151.4
|
TSR2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr22_+_39916558 | 0.01 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr6_-_160210715 | 0.01 |
ENST00000392168.2
ENST00000321394.7 |
TCP1
|
t-complex 1 |
| chr17_-_76713100 | 0.01 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr12_-_120241187 | 0.01 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr2_-_74618964 | 0.01 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr5_+_154393260 | 0.01 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr2_-_40680578 | 0.01 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_+_73882144 | 0.01 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr3_-_197024394 | 0.01 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_-_185538849 | 0.01 |
ENST00000421047.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_-_214016314 | 0.01 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr4_+_184020398 | 0.01 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chrX_+_128913906 | 0.01 |
ENST00000356892.3
|
SASH3
|
SAM and SH3 domain containing 3 |
| chr1_-_43855444 | 0.01 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr8_+_27632047 | 0.01 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr16_+_22019404 | 0.01 |
ENST00000542527.2
ENST00000569656.1 ENST00000562695.1 |
C16orf52
|
chromosome 16 open reading frame 52 |
| chr3_-_52713729 | 0.00 |
ENST00000296302.7
ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1
|
polybromo 1 |
| chr19_-_3029011 | 0.00 |
ENST00000590536.1
ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr15_+_80364901 | 0.00 |
ENST00000560228.1
ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr19_+_57640011 | 0.00 |
ENST00000598197.1
|
USP29
|
ubiquitin specific peptidase 29 |
| chr6_+_155443048 | 0.00 |
ENST00000535583.1
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr5_-_150138551 | 0.00 |
ENST00000446090.2
ENST00000447998.2 |
DCTN4
|
dynactin 4 (p62) |
| chr1_+_43855545 | 0.00 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr1_-_31538517 | 0.00 |
ENST00000440538.2
ENST00000423018.2 ENST00000424085.2 ENST00000426105.2 ENST00000257075.5 ENST00000373747.3 ENST00000525843.1 ENST00000373742.2 |
PUM1
|
pumilio RNA-binding family member 1 |
| chr13_-_41706864 | 0.00 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr8_+_35649365 | 0.00 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5B
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.2 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 0.6 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0032765 | lymphotoxin A production(GO:0032641) positive regulation of mast cell cytokine production(GO:0032765) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0031106 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0070433 | negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.2 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.0 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0001502 | cartilage condensation(GO:0001502) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.2 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.3 | GO:0005916 | fascia adherens(GO:0005916) catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.2 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |