Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ARID5A
Z-value: 0.75
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97202480_97202499 | -0.31 | 5.4e-01 | Click! |
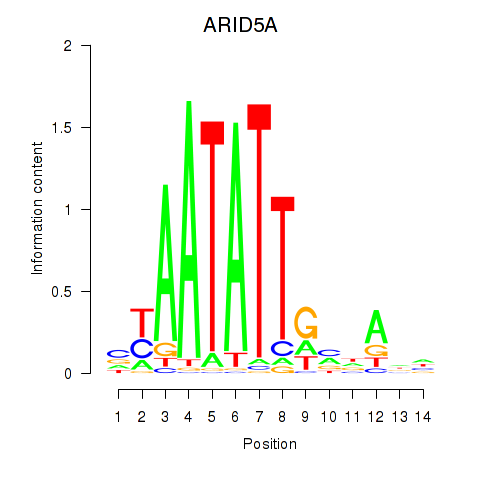
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_106959552 | 0.72 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883 |
| chr4_+_28437071 | 0.40 |
ENST00000509416.1
|
RP11-123O22.1
|
RP11-123O22.1 |
| chr2_+_152214098 | 0.40 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chrX_+_70521584 | 0.38 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr4_-_122148620 | 0.37 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr8_-_150563 | 0.34 |
ENST00000523795.2
|
RP11-585F1.10
|
Protein LOC100286914 |
| chr3_-_74570291 | 0.31 |
ENST00000263665.6
|
CNTN3
|
contactin 3 (plasmacytoma associated) |
| chr1_-_153013588 | 0.30 |
ENST00000360379.3
|
SPRR2D
|
small proline-rich protein 2D |
| chr1_-_100643765 | 0.29 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr2_-_98972468 | 0.28 |
ENST00000454230.1
|
AC092675.3
|
Uncharacterized protein |
| chr17_+_61473104 | 0.27 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr19_+_54135310 | 0.26 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr3_+_142342240 | 0.25 |
ENST00000497199.1
|
PLS1
|
plastin 1 |
| chr6_+_32605134 | 0.25 |
ENST00000343139.5
ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr7_-_92777606 | 0.25 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chrX_+_41583408 | 0.24 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr10_-_21661870 | 0.23 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr10_+_78078088 | 0.23 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr22_+_25615489 | 0.23 |
ENST00000398215.2
|
CRYBB2
|
crystallin, beta B2 |
| chr2_+_183982255 | 0.22 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr2_+_223725723 | 0.22 |
ENST00000535678.1
|
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr11_-_112034803 | 0.22 |
ENST00000528832.1
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr21_-_19858196 | 0.22 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr1_+_79115503 | 0.21 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr20_-_5426380 | 0.21 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr17_-_47925379 | 0.20 |
ENST00000352793.2
ENST00000334568.4 ENST00000398154.1 ENST00000436235.1 ENST00000326219.5 |
TAC4
|
tachykinin 4 (hemokinin) |
| chr3_-_107596910 | 0.19 |
ENST00000464359.2
ENST00000464823.1 ENST00000466155.1 ENST00000473528.2 ENST00000608306.1 ENST00000488852.1 ENST00000608137.1 ENST00000608307.1 ENST00000609429.1 ENST00000601385.1 ENST00000475362.1 ENST00000600240.1 ENST00000600749.1 |
LINC00635
|
long intergenic non-protein coding RNA 635 |
| chr5_+_61874562 | 0.19 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr19_-_58204128 | 0.19 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr6_-_52859046 | 0.19 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr2_-_228244013 | 0.19 |
ENST00000304568.3
|
TM4SF20
|
transmembrane 4 L six family member 20 |
| chr7_-_107883678 | 0.19 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr14_+_37641012 | 0.18 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr3_+_101546827 | 0.18 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr2_+_113321939 | 0.18 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr3_+_42885450 | 0.18 |
ENST00000492609.1
|
ACKR2
|
atypical chemokine receptor 2 |
| chr6_-_74161977 | 0.18 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr5_+_118690466 | 0.17 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr3_-_106959424 | 0.17 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr18_-_61329118 | 0.17 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr14_+_50291993 | 0.17 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr3_-_150421752 | 0.17 |
ENST00000498386.1
|
FAM194A
|
family with sequence similarity 194, member A |
| chr2_-_191115229 | 0.17 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr2_+_29353520 | 0.17 |
ENST00000438819.1
|
CLIP4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr12_-_48597170 | 0.16 |
ENST00000310248.2
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1 |
| chr7_-_16685422 | 0.16 |
ENST00000306999.2
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr1_+_41448820 | 0.16 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr6_+_107077471 | 0.15 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr1_+_212475148 | 0.15 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr1_+_76251912 | 0.15 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr5_+_147582348 | 0.15 |
ENST00000514389.1
|
SPINK6
|
serine peptidase inhibitor, Kazal type 6 |
| chr18_+_61554932 | 0.15 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr6_+_139135648 | 0.15 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr18_+_68002675 | 0.15 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr4_-_48683188 | 0.14 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr3_+_142342228 | 0.14 |
ENST00000337777.3
|
PLS1
|
plastin 1 |
| chr6_+_30035307 | 0.14 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr2_+_201981663 | 0.14 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_-_119869846 | 0.14 |
ENST00000457719.1
|
RP11-418J17.3
|
RP11-418J17.3 |
| chr20_-_55100981 | 0.14 |
ENST00000243913.4
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr6_-_136571400 | 0.14 |
ENST00000418509.2
ENST00000420702.1 ENST00000451457.2 |
MTFR2
|
mitochondrial fission regulator 2 |
| chr9_-_75653627 | 0.14 |
ENST00000446946.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr2_+_183982238 | 0.14 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr17_+_35767301 | 0.13 |
ENST00000225396.6
ENST00000417170.1 ENST00000590005.1 ENST00000590957.1 |
TADA2A
|
transcriptional adaptor 2A |
| chr19_-_35264089 | 0.13 |
ENST00000588760.1
ENST00000329285.8 ENST00000587354.2 |
ZNF599
|
zinc finger protein 599 |
| chr10_-_104866395 | 0.13 |
ENST00000458345.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr17_+_68047418 | 0.13 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr19_+_35417798 | 0.12 |
ENST00000303586.7
ENST00000439785.1 ENST00000601540.1 |
ZNF30
|
zinc finger protein 30 |
| chr1_+_163039143 | 0.12 |
ENST00000531057.1
ENST00000527809.1 ENST00000367908.4 |
RGS4
|
regulator of G-protein signaling 4 |
| chr4_+_76871883 | 0.12 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr10_-_15902449 | 0.12 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr17_+_57233087 | 0.12 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chrX_+_107288239 | 0.12 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr15_-_59981479 | 0.11 |
ENST00000607373.1
|
BNIP2
|
BCL2/adenovirus E1B 19kDa interacting protein 2 |
| chr17_-_71223839 | 0.11 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr12_-_133787772 | 0.11 |
ENST00000545350.1
|
AC226150.4
|
Uncharacterized protein |
| chr8_+_67104323 | 0.11 |
ENST00000518412.1
ENST00000518035.1 ENST00000517725.1 |
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr8_-_66701319 | 0.11 |
ENST00000379419.4
|
PDE7A
|
phosphodiesterase 7A |
| chr16_-_3422283 | 0.11 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr7_+_16685756 | 0.11 |
ENST00000415365.1
ENST00000258761.3 ENST00000433922.2 ENST00000452975.2 ENST00000405202.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr11_+_69061594 | 0.11 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr7_+_138943265 | 0.11 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr3_-_69171708 | 0.11 |
ENST00000420581.2
|
LMOD3
|
leiomodin 3 (fetal) |
| chr14_+_56584414 | 0.10 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_+_102290772 | 0.10 |
ENST00000507450.1
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chrX_+_153991025 | 0.10 |
ENST00000369550.5
|
DKC1
|
dyskeratosis congenita 1, dyskerin |
| chr18_-_68004529 | 0.10 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chrX_+_107288197 | 0.10 |
ENST00000415430.3
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr1_-_89736434 | 0.10 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr6_+_10528560 | 0.10 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr15_-_59949667 | 0.10 |
ENST00000396061.1
|
GTF2A2
|
general transcription factor IIA, 2, 12kDa |
| chr20_-_52612705 | 0.10 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr11_-_2170786 | 0.09 |
ENST00000300632.5
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr3_-_150421728 | 0.09 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr1_-_156722195 | 0.09 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr6_-_133055896 | 0.09 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr20_+_37590942 | 0.09 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr14_+_90722839 | 0.09 |
ENST00000261303.8
ENST00000553835.1 |
PSMC1
|
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr7_-_45957011 | 0.09 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr8_-_37411648 | 0.09 |
ENST00000519738.1
|
RP11-150O12.1
|
RP11-150O12.1 |
| chr4_+_141264597 | 0.09 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr18_-_53089723 | 0.09 |
ENST00000561992.1
ENST00000562512.2 |
TCF4
|
transcription factor 4 |
| chr4_+_69962185 | 0.09 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_-_3195981 | 0.09 |
ENST00000425384.2
ENST00000435043.2 |
RP1-40E16.9
|
RP1-40E16.9 |
| chr1_+_104159999 | 0.09 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr19_-_9879293 | 0.09 |
ENST00000397902.2
ENST00000592859.1 ENST00000588267.1 |
ZNF846
|
zinc finger protein 846 |
| chr11_+_61976137 | 0.09 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr4_-_95264008 | 0.08 |
ENST00000295256.5
|
HPGDS
|
hematopoietic prostaglandin D synthase |
| chr6_-_18249971 | 0.08 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr6_+_32605195 | 0.08 |
ENST00000374949.2
|
HLA-DQA1
|
major histocompatibility complex, class II, DQ alpha 1 |
| chr6_-_26235206 | 0.08 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr3_+_101280677 | 0.08 |
ENST00000309922.6
ENST00000495642.1 |
TRMT10C
|
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr1_+_76251879 | 0.08 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr4_-_110624564 | 0.08 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr8_+_128426535 | 0.08 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr10_-_63995871 | 0.08 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr5_-_43397184 | 0.08 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr22_-_23922448 | 0.08 |
ENST00000438703.1
ENST00000330377.2 |
IGLL1
|
immunoglobulin lambda-like polypeptide 1 |
| chr12_-_49582978 | 0.08 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chrX_-_100662881 | 0.08 |
ENST00000218516.3
|
GLA
|
galactosidase, alpha |
| chr21_+_30673091 | 0.08 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr19_+_35417844 | 0.08 |
ENST00000601957.1
|
ZNF30
|
zinc finger protein 30 |
| chr4_-_103749105 | 0.08 |
ENST00000394801.4
ENST00000394804.2 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_116654376 | 0.08 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr12_-_48099754 | 0.08 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr7_-_92157747 | 0.08 |
ENST00000428214.1
ENST00000438045.1 |
PEX1
|
peroxisomal biogenesis factor 1 |
| chr9_+_70856899 | 0.08 |
ENST00000377342.5
ENST00000478048.1 |
CBWD3
|
COBW domain containing 3 |
| chr10_-_61720640 | 0.08 |
ENST00000521074.1
ENST00000444900.1 |
C10orf40
|
chromosome 10 open reading frame 40 |
| chrX_+_23682379 | 0.08 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr13_-_52703187 | 0.08 |
ENST00000355568.4
|
NEK5
|
NIMA-related kinase 5 |
| chr1_+_144989309 | 0.08 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr6_-_130543958 | 0.08 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr12_+_10460417 | 0.08 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr2_+_242312264 | 0.08 |
ENST00000445489.1
|
FARP2
|
FERM, RhoGEF and pleckstrin domain protein 2 |
| chr3_-_142297668 | 0.08 |
ENST00000350721.4
ENST00000383101.3 |
ATR
|
ataxia telangiectasia and Rad3 related |
| chrX_-_118699325 | 0.08 |
ENST00000320339.4
ENST00000371594.4 ENST00000536133.1 |
CXorf56
|
chromosome X open reading frame 56 |
| chr7_-_92157760 | 0.08 |
ENST00000248633.4
|
PEX1
|
peroxisomal biogenesis factor 1 |
| chr9_+_112542591 | 0.07 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr12_-_106480587 | 0.07 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chr16_+_55542910 | 0.07 |
ENST00000262134.5
|
LPCAT2
|
lysophosphatidylcholine acyltransferase 2 |
| chr12_+_60083118 | 0.07 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_125366018 | 0.07 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr3_-_69129501 | 0.07 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr4_-_71532207 | 0.07 |
ENST00000543780.1
ENST00000391614.3 |
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_13910194 | 0.07 |
ENST00000376057.4
ENST00000510906.1 |
PDPN
|
podoplanin |
| chr4_+_55095428 | 0.07 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_+_14716606 | 0.07 |
ENST00000253697.3
ENST00000435614.1 ENST00000412910.1 |
C3orf20
|
chromosome 3 open reading frame 20 |
| chr2_-_225362533 | 0.07 |
ENST00000451538.1
|
CUL3
|
cullin 3 |
| chr3_-_131221790 | 0.07 |
ENST00000512877.1
ENST00000264995.3 ENST00000511168.1 ENST00000425847.2 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr2_-_10587897 | 0.07 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr11_+_17316870 | 0.07 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr6_+_151561506 | 0.07 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_+_66797687 | 0.07 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr19_+_33865218 | 0.07 |
ENST00000585933.2
|
CEBPG
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr4_-_69111401 | 0.07 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr9_-_13175823 | 0.07 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr7_-_45956856 | 0.07 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr3_-_131221771 | 0.07 |
ENST00000510154.1
ENST00000507669.1 |
MRPL3
|
mitochondrial ribosomal protein L3 |
| chr6_-_33048503 | 0.07 |
ENST00000453337.1
ENST00000417724.1 |
HLA-DPA1
|
major histocompatibility complex, class II, DP alpha 1 |
| chr16_-_58328923 | 0.07 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr19_-_20748614 | 0.07 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr8_+_92261516 | 0.07 |
ENST00000276609.3
ENST00000309536.2 |
SLC26A7
|
solute carrier family 26 (anion exchanger), member 7 |
| chr7_-_22233442 | 0.07 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_+_100737954 | 0.07 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr3_+_11196206 | 0.07 |
ENST00000431010.2
|
HRH1
|
histamine receptor H1 |
| chr1_-_154600421 | 0.06 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr20_+_5986727 | 0.06 |
ENST00000378863.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr6_+_151561085 | 0.06 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr7_+_116593536 | 0.06 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr5_+_59783540 | 0.06 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr10_+_127661942 | 0.06 |
ENST00000417114.1
ENST00000445510.1 ENST00000368691.1 |
FANK1
|
fibronectin type III and ankyrin repeat domains 1 |
| chr12_-_48099773 | 0.06 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr10_+_114710516 | 0.06 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_158901329 | 0.06 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr4_+_71019903 | 0.06 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr2_+_219472488 | 0.06 |
ENST00000450993.2
|
PLCD4
|
phospholipase C, delta 4 |
| chr2_-_133429091 | 0.06 |
ENST00000345008.6
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr5_+_128795958 | 0.06 |
ENST00000274487.4
|
ADAMTS19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
| chr16_+_10479906 | 0.06 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr7_-_24797546 | 0.06 |
ENST00000414428.1
ENST00000419307.1 ENST00000342947.3 |
DFNA5
|
deafness, autosomal dominant 5 |
| chr5_+_64064748 | 0.06 |
ENST00000381070.3
ENST00000508024.1 |
CWC27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr5_+_140602904 | 0.06 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr1_+_241695424 | 0.06 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr1_-_149459549 | 0.06 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr22_-_38699003 | 0.06 |
ENST00000451964.1
|
CSNK1E
|
casein kinase 1, epsilon |
| chr14_-_92198403 | 0.06 |
ENST00000553329.1
ENST00000256343.3 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr16_-_90128733 | 0.06 |
ENST00000325921.6
|
PRDM7
|
PR domain containing 7 |
| chr19_+_52076425 | 0.06 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr13_-_41837620 | 0.06 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr14_+_58797974 | 0.06 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr14_-_92247032 | 0.06 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr15_-_78913628 | 0.06 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr12_+_4758264 | 0.06 |
ENST00000266544.5
|
NDUFA9
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr12_-_85430024 | 0.05 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr18_-_50240 | 0.05 |
ENST00000573909.1
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr2_+_114195268 | 0.05 |
ENST00000259199.4
ENST00000416503.2 ENST00000433343.2 |
CBWD2
|
COBW domain containing 2 |
| chr10_-_102027420 | 0.05 |
ENST00000354105.4
|
CWF19L1
|
CWF19-like 1, cell cycle control (S. pombe) |
| chr11_-_26588634 | 0.05 |
ENST00000436318.2
ENST00000281268.8 |
MUC15
|
mucin 15, cell surface associated |
| chr2_+_157330081 | 0.05 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr18_+_52258390 | 0.05 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.1 | 0.4 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.2 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0060152 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.2 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.0 | 0.1 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.2 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.0 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:1904882 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.0 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.0 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |