Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
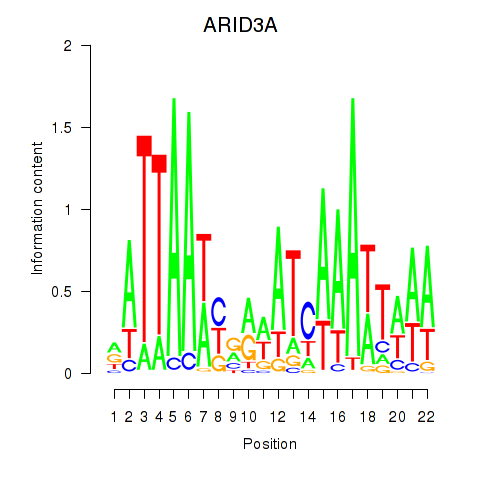
Results for ARID3A
Z-value: 0.85
Transcription factors associated with ARID3A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID3A
|
ENSG00000116017.6 | AT-rich interaction domain 3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID3A | hg19_v2_chr19_+_926000_926046 | 0.24 | 6.4e-01 | Click! |
Activity profile of ARID3A motif
Sorted Z-values of ARID3A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_74606223 | 1.05 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr12_+_113344582 | 0.76 |
ENST00000202917.5
ENST00000445409.2 ENST00000452357.2 |
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_-_165424973 | 0.55 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr5_-_150467221 | 0.45 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr18_+_61554932 | 0.44 |
ENST00000299502.4
ENST00000457692.1 ENST00000413956.1 |
SERPINB2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr11_+_18154059 | 0.43 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr3_-_169487617 | 0.36 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr14_+_37641012 | 0.32 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr9_-_110540419 | 0.32 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr2_-_191115229 | 0.29 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr18_+_29671812 | 0.28 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr20_+_55099542 | 0.28 |
ENST00000371328.3
|
FAM209A
|
family with sequence similarity 209, member A |
| chr8_+_39759794 | 0.27 |
ENST00000518804.1
ENST00000519154.1 ENST00000522495.1 ENST00000522840.1 |
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr12_-_12714006 | 0.26 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr11_-_327537 | 0.26 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr7_-_139763521 | 0.26 |
ENST00000263549.3
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr12_+_69186125 | 0.26 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr4_+_70916119 | 0.25 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr1_-_183538319 | 0.24 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr4_-_48683188 | 0.24 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr4_+_69917078 | 0.24 |
ENST00000502942.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr7_+_16828866 | 0.23 |
ENST00000597084.1
|
AC073333.1
|
Uncharacterized protein |
| chr5_+_159656437 | 0.23 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr19_-_48614063 | 0.22 |
ENST00000599921.1
ENST00000599111.1 |
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr6_-_108278456 | 0.22 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr2_+_183982255 | 0.22 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr12_-_12714025 | 0.22 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr3_+_138340067 | 0.22 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr1_+_50459990 | 0.21 |
ENST00000448346.1
|
AL645730.2
|
AL645730.2 |
| chr10_-_69597828 | 0.21 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr16_-_2770216 | 0.20 |
ENST00000302641.3
|
PRSS27
|
protease, serine 27 |
| chr10_-_90611566 | 0.20 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr17_-_71223839 | 0.20 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr3_-_130745403 | 0.19 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr17_-_12920907 | 0.18 |
ENST00000609757.1
ENST00000581499.2 ENST00000580504.1 |
ELAC2
|
elaC ribonuclease Z 2 |
| chrX_+_41583408 | 0.18 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr5_+_68513622 | 0.18 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr2_+_234294585 | 0.18 |
ENST00000447484.1
|
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr3_+_119298280 | 0.18 |
ENST00000481816.1
|
ADPRH
|
ADP-ribosylarginine hydrolase |
| chr1_-_39395165 | 0.17 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chrX_-_20159934 | 0.17 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr4_-_68749745 | 0.17 |
ENST00000283916.6
|
TMPRSS11D
|
transmembrane protease, serine 11D |
| chr16_+_14802801 | 0.17 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr17_-_12920994 | 0.17 |
ENST00000609101.1
|
ELAC2
|
elaC ribonuclease Z 2 |
| chr21_-_22175341 | 0.17 |
ENST00000416768.1
ENST00000452561.1 ENST00000419299.1 ENST00000437238.1 |
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr6_+_3259148 | 0.17 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr7_+_64838712 | 0.16 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr12_+_97306295 | 0.16 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr7_+_64838786 | 0.16 |
ENST00000450302.2
|
ZNF92
|
zinc finger protein 92 |
| chr7_-_22539771 | 0.16 |
ENST00000406890.2
ENST00000424363.1 |
STEAP1B
|
STEAP family member 1B |
| chr16_-_31564881 | 0.16 |
ENST00000567765.1
|
CTD-2014E2.6
|
CTD-2014E2.6 |
| chr3_+_169629354 | 0.16 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr14_+_39734482 | 0.16 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr2_+_208414985 | 0.15 |
ENST00000414681.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr16_-_30064244 | 0.15 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr1_+_41448820 | 0.15 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr4_-_80994619 | 0.15 |
ENST00000404191.1
|
ANTXR2
|
anthrax toxin receptor 2 |
| chr17_+_41006095 | 0.14 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase, copper containing 3 |
| chr14_+_101295638 | 0.14 |
ENST00000523671.2
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr2_+_181988560 | 0.14 |
ENST00000424170.1
ENST00000435411.1 |
AC104820.2
|
AC104820.2 |
| chr15_-_99057551 | 0.14 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr7_+_76090993 | 0.14 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr1_-_205391178 | 0.14 |
ENST00000367153.4
ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1
|
LEM domain containing 1 |
| chr11_+_111788738 | 0.13 |
ENST00000529342.1
|
C11orf52
|
chromosome 11 open reading frame 52 |
| chrX_+_21857717 | 0.13 |
ENST00000379484.5
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2 |
| chr6_+_3259122 | 0.13 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr7_+_89783689 | 0.13 |
ENST00000297205.2
|
STEAP1
|
six transmembrane epithelial antigen of the prostate 1 |
| chr12_+_32832134 | 0.13 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr10_+_91092241 | 0.13 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr20_+_31823091 | 0.13 |
ENST00000601172.1
|
AL121901.1
|
Nasopharyngeal carcinoma-related protein YH1; Uncharacterized protein |
| chr21_-_43735628 | 0.13 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr14_-_53162361 | 0.13 |
ENST00000395686.3
|
ERO1L
|
ERO1-like (S. cerevisiae) |
| chr8_-_2585929 | 0.13 |
ENST00000519393.1
ENST00000520842.1 ENST00000520570.1 ENST00000517357.1 ENST00000517984.1 ENST00000523971.1 |
RP11-134O21.1
|
RP11-134O21.1 |
| chr13_-_30951282 | 0.13 |
ENST00000420219.1
|
LINC00426
|
long intergenic non-protein coding RNA 426 |
| chr8_+_74903580 | 0.13 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr3_-_121379739 | 0.13 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr13_-_33780133 | 0.13 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr4_-_80994210 | 0.13 |
ENST00000403729.2
|
ANTXR2
|
anthrax toxin receptor 2 |
| chrX_-_102531717 | 0.13 |
ENST00000372680.1
|
TCEAL5
|
transcription elongation factor A (SII)-like 5 |
| chr4_+_113568207 | 0.13 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr3_+_118892411 | 0.12 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr19_-_46145696 | 0.12 |
ENST00000588172.1
|
EML2
|
echinoderm microtubule associated protein like 2 |
| chrX_-_114252193 | 0.12 |
ENST00000243213.1
|
IL13RA2
|
interleukin 13 receptor, alpha 2 |
| chr6_-_138833630 | 0.12 |
ENST00000533765.1
|
NHSL1
|
NHS-like 1 |
| chr12_+_60083118 | 0.12 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_+_327171 | 0.12 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr11_+_17316870 | 0.12 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr12_-_21910775 | 0.12 |
ENST00000539782.1
|
LDHB
|
lactate dehydrogenase B |
| chr7_+_138943265 | 0.12 |
ENST00000483726.1
|
UBN2
|
ubinuclein 2 |
| chr17_+_18684563 | 0.12 |
ENST00000476139.1
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr16_+_84801852 | 0.12 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr21_+_45209394 | 0.12 |
ENST00000497547.1
|
RRP1
|
ribosomal RNA processing 1 |
| chr4_+_141264597 | 0.11 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr1_-_152539248 | 0.11 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr4_-_169239921 | 0.11 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr8_+_67687413 | 0.11 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr5_-_43557791 | 0.11 |
ENST00000338972.4
ENST00000511321.1 ENST00000515338.1 |
PAIP1
|
poly(A) binding protein interacting protein 1 |
| chr1_-_235098861 | 0.11 |
ENST00000458044.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chr19_+_37742773 | 0.11 |
ENST00000438770.2
ENST00000591116.1 ENST00000592712.1 |
AC012309.5
|
AC012309.5 |
| chr20_+_3767547 | 0.11 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr8_-_23282797 | 0.11 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr7_+_65552756 | 0.11 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr19_-_48614033 | 0.11 |
ENST00000354276.3
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr6_-_100016527 | 0.11 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr12_+_96883347 | 0.10 |
ENST00000524981.4
ENST00000298953.3 |
C12orf55
|
chromosome 12 open reading frame 55 |
| chr3_+_138340049 | 0.10 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr4_+_74301880 | 0.10 |
ENST00000395792.2
ENST00000226359.2 |
AFP
|
alpha-fetoprotein |
| chr7_+_93690435 | 0.10 |
ENST00000438538.1
|
AC003092.1
|
AC003092.1 |
| chr17_-_15554940 | 0.10 |
ENST00000455584.2
|
RP11-385D13.1
|
Uncharacterized protein |
| chr19_+_11200038 | 0.10 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr8_+_23104130 | 0.10 |
ENST00000313219.7
ENST00000519984.1 |
CHMP7
|
charged multivesicular body protein 7 |
| chr16_-_72128270 | 0.10 |
ENST00000426362.2
|
TXNL4B
|
thioredoxin-like 4B |
| chr17_-_76824975 | 0.10 |
ENST00000586066.2
|
USP36
|
ubiquitin specific peptidase 36 |
| chr12_+_32832203 | 0.09 |
ENST00000553257.1
ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L
|
dynamin 1-like |
| chr4_+_78829479 | 0.09 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr19_-_20748541 | 0.09 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr6_+_151561085 | 0.09 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr17_-_41050716 | 0.09 |
ENST00000417193.1
ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671
|
long intergenic non-protein coding RNA 671 |
| chr19_+_49199209 | 0.09 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr3_-_139396853 | 0.09 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr14_+_97263641 | 0.09 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr10_-_115904361 | 0.09 |
ENST00000428953.1
ENST00000543782.1 |
C10orf118
|
chromosome 10 open reading frame 118 |
| chr8_+_86099884 | 0.09 |
ENST00000517476.1
ENST00000521429.1 |
E2F5
|
E2F transcription factor 5, p130-binding |
| chr2_-_176867501 | 0.09 |
ENST00000535310.1
|
KIAA1715
|
KIAA1715 |
| chr7_-_66460563 | 0.09 |
ENST00000246868.2
|
SBDS
|
Shwachman-Bodian-Diamond syndrome |
| chr3_+_167582561 | 0.09 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr19_-_9896672 | 0.09 |
ENST00000589412.1
ENST00000586814.1 |
ZNF846
|
zinc finger protein 846 |
| chr8_-_23282820 | 0.09 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr1_+_224544572 | 0.09 |
ENST00000366857.5
ENST00000366856.3 |
CNIH4
|
cornichon family AMPA receptor auxiliary protein 4 |
| chr14_+_96039882 | 0.09 |
ENST00000556346.1
ENST00000553785.1 |
RP11-1070N10.4
|
RP11-1070N10.4 |
| chr2_+_73429386 | 0.08 |
ENST00000398468.3
|
NOTO
|
notochord homeobox |
| chr6_-_46703430 | 0.08 |
ENST00000537365.1
|
PLA2G7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr3_+_12329358 | 0.08 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr13_-_20110902 | 0.08 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr19_+_36239576 | 0.08 |
ENST00000587751.1
|
LIN37
|
lin-37 homolog (C. elegans) |
| chr8_-_104153703 | 0.08 |
ENST00000521246.1
|
C8orf56
|
chromosome 8 open reading frame 56 |
| chr1_+_207943667 | 0.08 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr2_+_183982238 | 0.08 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chr2_+_119699742 | 0.08 |
ENST00000327097.4
|
MARCO
|
macrophage receptor with collagenous structure |
| chr22_-_32767017 | 0.08 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr2_-_11606275 | 0.08 |
ENST00000381525.3
ENST00000362009.4 |
E2F6
|
E2F transcription factor 6 |
| chr7_-_6048650 | 0.08 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr1_-_247267580 | 0.08 |
ENST00000366501.1
ENST00000366500.1 ENST00000476158.2 ENST00000448299.2 ENST00000358785.4 ENST00000343381.6 |
ZNF669
|
zinc finger protein 669 |
| chr19_-_39826639 | 0.07 |
ENST00000602185.1
ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG
|
glia maturation factor, gamma |
| chr11_+_129936836 | 0.07 |
ENST00000597197.1
|
AP003041.2
|
Uncharacterized protein |
| chr5_+_141346385 | 0.07 |
ENST00000513019.1
ENST00000356143.1 |
RNF14
|
ring finger protein 14 |
| chr2_+_90248739 | 0.07 |
ENST00000468879.1
|
IGKV1D-43
|
immunoglobulin kappa variable 1D-43 |
| chr11_-_70507901 | 0.07 |
ENST00000449833.2
ENST00000357171.3 ENST00000449116.2 |
SHANK2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr6_-_64029879 | 0.07 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr7_+_107224364 | 0.07 |
ENST00000491150.1
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr1_-_180991978 | 0.07 |
ENST00000542060.1
ENST00000258301.5 |
STX6
|
syntaxin 6 |
| chr8_+_67687773 | 0.07 |
ENST00000518388.1
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr17_+_75181292 | 0.07 |
ENST00000431431.2
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr19_+_107471 | 0.07 |
ENST00000585993.1
|
OR4F17
|
olfactory receptor, family 4, subfamily F, member 17 |
| chr3_+_118892362 | 0.07 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr6_+_24403144 | 0.07 |
ENST00000274747.7
ENST00000543597.1 ENST00000535061.1 ENST00000378353.1 ENST00000378386.3 ENST00000443868.2 |
MRS2
|
MRS2 magnesium transporter |
| chr2_+_20101786 | 0.07 |
ENST00000607190.1
|
RP11-79O8.1
|
RP11-79O8.1 |
| chr3_+_57875738 | 0.07 |
ENST00000417128.1
ENST00000438794.1 |
SLMAP
|
sarcolemma associated protein |
| chr4_+_71017791 | 0.07 |
ENST00000502294.1
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr21_-_23058648 | 0.07 |
ENST00000416182.1
|
AF241725.6
|
AF241725.6 |
| chr8_-_74495065 | 0.07 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr19_-_38183228 | 0.07 |
ENST00000587199.1
|
ZFP30
|
ZFP30 zinc finger protein |
| chr21_-_22175450 | 0.06 |
ENST00000435279.2
|
LINC00320
|
long intergenic non-protein coding RNA 320 |
| chr1_+_234509413 | 0.06 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr1_-_154600421 | 0.06 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr2_+_210443993 | 0.06 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr12_+_122880045 | 0.06 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr16_+_30064444 | 0.06 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_119699864 | 0.06 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr4_+_100737954 | 0.06 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr6_-_116381918 | 0.06 |
ENST00000606080.1
|
FRK
|
fyn-related kinase |
| chr8_+_32579271 | 0.06 |
ENST00000518084.1
|
NRG1
|
neuregulin 1 |
| chr20_-_60573188 | 0.06 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr17_+_7531281 | 0.06 |
ENST00000575729.1
ENST00000340624.5 |
SHBG
|
sex hormone-binding globulin |
| chr1_-_227505289 | 0.06 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_+_186158169 | 0.06 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr2_+_110551851 | 0.06 |
ENST00000272454.6
ENST00000430736.1 ENST00000016946.3 ENST00000441344.1 |
RGPD5
|
RANBP2-like and GRIP domain containing 5 |
| chr11_-_92930556 | 0.06 |
ENST00000529184.1
|
SLC36A4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr2_+_102928009 | 0.06 |
ENST00000404917.2
ENST00000447231.1 |
IL1RL1
|
interleukin 1 receptor-like 1 |
| chr10_-_11574274 | 0.06 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr3_-_139396801 | 0.06 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr10_+_97733786 | 0.06 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr6_-_13328050 | 0.06 |
ENST00000420456.1
|
TBC1D7
|
TBC1 domain family, member 7 |
| chr1_-_151162606 | 0.06 |
ENST00000354473.4
ENST00000368892.4 |
VPS72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr5_-_110062384 | 0.06 |
ENST00000429839.2
|
TMEM232
|
transmembrane protein 232 |
| chr4_-_69536346 | 0.06 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr16_-_46649905 | 0.06 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr7_+_156902674 | 0.06 |
ENST00000594086.1
|
AC006967.1
|
Protein LOC100996426 |
| chr22_-_32766972 | 0.06 |
ENST00000382084.4
ENST00000382086.2 |
RFPL3S
|
RFPL3 antisense |
| chr5_-_16916624 | 0.06 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr16_+_30064411 | 0.06 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr9_-_69229650 | 0.06 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr1_+_207002222 | 0.06 |
ENST00000270218.6
|
IL19
|
interleukin 19 |
| chr19_+_13001840 | 0.06 |
ENST00000222214.5
ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH
|
glutaryl-CoA dehydrogenase |
| chr6_-_32977345 | 0.05 |
ENST00000450833.2
ENST00000374813.1 ENST00000229829.5 |
HLA-DOA
|
major histocompatibility complex, class II, DO alpha |
| chr17_+_71228740 | 0.05 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr8_+_18248786 | 0.05 |
ENST00000520116.1
|
NAT2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr17_-_71228357 | 0.05 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr13_-_88323218 | 0.05 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr2_+_183582774 | 0.05 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID3A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.3 | GO:0072684 | mitochondrial tRNA 3'-trailer cleavage, endonucleolytic(GO:0072684) |
| 0.1 | 0.5 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.2 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.1 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.0 | GO:0034698 | response to follicle-stimulating hormone(GO:0032354) response to gonadotropin(GO:0034698) |
| 0.0 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:1903923 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.3 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.2 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.0 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.0 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |