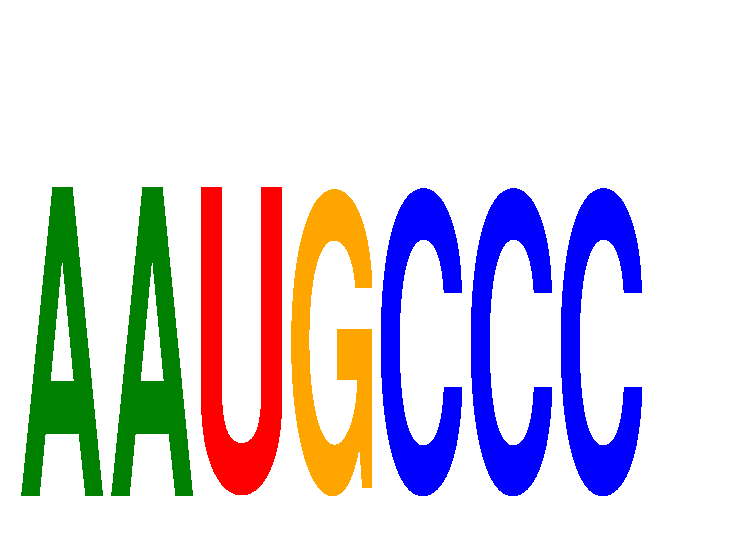Project
NHBE cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.33
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-365a-3p
|
MIMAT0000710 |
|
hsa-miR-365b-3p
|
MIMAT0022834 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_123603137 | 0.13 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr3_+_23986748 | 0.13 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr2_+_46926048 | 0.09 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr1_-_46598284 | 0.08 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr11_+_34642656 | 0.07 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr13_-_46425865 | 0.07 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr10_-_98273668 | 0.07 |
ENST00000357947.3
|
TLL2
|
tolloid-like 2 |
| chr5_-_142065612 | 0.07 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr1_-_91487013 | 0.06 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr1_-_244013384 | 0.06 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr12_-_42538657 | 0.06 |
ENST00000398675.3
|
GXYLT1
|
glucoside xylosyltransferase 1 |
| chr4_-_103748880 | 0.06 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_-_48072574 | 0.06 |
ENST00000434704.2
|
DLX3
|
distal-less homeobox 3 |
| chr20_+_56884752 | 0.05 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr2_-_183903133 | 0.05 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr10_-_98346801 | 0.05 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr2_-_153574480 | 0.05 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_-_78225482 | 0.05 |
ENST00000524778.1
ENST00000370794.3 ENST00000370793.1 ENST00000370792.3 |
USP33
|
ubiquitin specific peptidase 33 |
| chr13_-_21476900 | 0.05 |
ENST00000400602.2
ENST00000255305.6 |
XPO4
|
exportin 4 |
| chr3_+_113775576 | 0.04 |
ENST00000485050.1
ENST00000281273.4 |
QTRTD1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr10_-_62149433 | 0.04 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_-_33481835 | 0.04 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr6_-_111804393 | 0.04 |
ENST00000368802.3
ENST00000368805.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr15_+_52121822 | 0.04 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr1_+_78470530 | 0.04 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chrX_-_153979315 | 0.04 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr5_-_90679145 | 0.04 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr6_-_46138676 | 0.04 |
ENST00000371383.2
ENST00000230565.3 |
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) |
| chr4_+_91048706 | 0.04 |
ENST00000509176.1
|
CCSER1
|
coiled-coil serine-rich protein 1 |
| chr5_+_172483347 | 0.04 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
| chr1_-_23857698 | 0.04 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr10_+_89622870 | 0.04 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chr4_+_38665810 | 0.04 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr6_+_16129308 | 0.04 |
ENST00000356840.3
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chrX_-_109561294 | 0.04 |
ENST00000372059.2
ENST00000262844.5 |
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr1_+_97187318 | 0.04 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr17_+_55333876 | 0.04 |
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2 |
| chr12_+_68042495 | 0.03 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr5_-_89825328 | 0.03 |
ENST00000500869.2
ENST00000315948.6 ENST00000509384.1 |
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr9_+_114659046 | 0.03 |
ENST00000374279.3
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr8_+_59323823 | 0.03 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr2_+_61108650 | 0.03 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr1_+_36396677 | 0.03 |
ENST00000373191.4
ENST00000397828.2 |
AGO3
|
argonaute RISC catalytic component 3 |
| chr14_-_61190754 | 0.03 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr12_-_75905374 | 0.03 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr1_-_155881156 | 0.03 |
ENST00000539040.1
ENST00000368323.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr1_-_22109682 | 0.03 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr16_-_79634595 | 0.03 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr8_-_38239732 | 0.03 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr5_+_112312416 | 0.03 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr21_+_38071430 | 0.03 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr16_+_53088885 | 0.03 |
ENST00000566029.1
ENST00000447540.1 |
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr12_+_56401268 | 0.03 |
ENST00000262032.5
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr12_+_66217911 | 0.03 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr19_-_49576198 | 0.03 |
ENST00000221444.1
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr8_+_124780672 | 0.03 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr14_-_55878538 | 0.03 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr2_-_180129484 | 0.03 |
ENST00000428443.3
|
SESTD1
|
SEC14 and spectrin domains 1 |
| chr5_-_39074479 | 0.02 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr13_-_53024725 | 0.02 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr3_-_113465065 | 0.02 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr2_-_208634287 | 0.02 |
ENST00000295417.3
|
FZD5
|
frizzled family receptor 5 |
| chr1_+_145611010 | 0.02 |
ENST00000369291.5
|
RNF115
|
ring finger protein 115 |
| chr15_+_96873921 | 0.02 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_-_82598511 | 0.02 |
ENST00000449740.2
ENST00000311489.4 ENST00000521360.1 ENST00000519964.1 ENST00000518202.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr4_+_37245799 | 0.02 |
ENST00000309447.5
|
KIAA1239
|
KIAA1239 |
| chr17_-_16118835 | 0.02 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr15_-_64995399 | 0.02 |
ENST00000559753.1
ENST00000560258.2 ENST00000559912.2 ENST00000326005.6 |
OAZ2
|
ornithine decarboxylase antizyme 2 |
| chr11_-_77532050 | 0.02 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr6_-_88411911 | 0.02 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr15_+_41523335 | 0.02 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr3_+_101292939 | 0.02 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr9_-_6007787 | 0.02 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr16_+_67063036 | 0.02 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chrX_-_3631635 | 0.02 |
ENST00000262848.5
|
PRKX
|
protein kinase, X-linked |
| chr10_+_180987 | 0.02 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr5_+_122181184 | 0.02 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr10_+_22610124 | 0.02 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr12_-_89918522 | 0.02 |
ENST00000529983.2
|
GALNT4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) |
| chr5_-_142783175 | 0.02 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr3_+_152552685 | 0.02 |
ENST00000305097.3
|
P2RY1
|
purinergic receptor P2Y, G-protein coupled, 1 |
| chr13_+_93879085 | 0.02 |
ENST00000377047.4
|
GPC6
|
glypican 6 |
| chr12_-_89919965 | 0.02 |
ENST00000548729.1
|
POC1B-GALNT4
|
POC1B-GALNT4 readthrough |
| chr2_+_179345173 | 0.02 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr17_+_68165657 | 0.02 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr21_-_28217721 | 0.02 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_+_43328004 | 0.01 |
ENST00000454177.1
ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK
|
SNF related kinase |
| chr13_+_114238997 | 0.01 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr11_+_2466218 | 0.01 |
ENST00000155840.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr11_-_27528301 | 0.01 |
ENST00000524596.1
ENST00000278193.2 |
LIN7C
|
lin-7 homolog C (C. elegans) |
| chr12_-_15942309 | 0.01 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr3_+_38495333 | 0.01 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr1_-_28415204 | 0.01 |
ENST00000373871.3
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr11_-_31839488 | 0.01 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chr3_-_48885228 | 0.01 |
ENST00000454963.1
ENST00000296446.8 ENST00000419216.1 ENST00000265563.8 |
PRKAR2A
|
protein kinase, cAMP-dependent, regulatory, type II, alpha |
| chr17_-_37557846 | 0.01 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr11_+_64073699 | 0.01 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr3_-_11762202 | 0.01 |
ENST00000445411.1
ENST00000404339.1 ENST00000273038.3 |
VGLL4
|
vestigial like 4 (Drosophila) |
| chr2_-_157189180 | 0.01 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_+_104654623 | 0.01 |
ENST00000311117.3
ENST00000334877.4 ENST00000257745.4 ENST00000334914.7 ENST00000478990.1 ENST00000495267.1 ENST00000476671.1 |
KMT2E
|
lysine (K)-specific methyltransferase 2E |
| chr1_-_205719295 | 0.01 |
ENST00000367142.4
|
NUCKS1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr17_+_28804380 | 0.01 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr10_-_120514720 | 0.01 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr4_-_11431389 | 0.01 |
ENST00000002596.5
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr2_-_39348137 | 0.01 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr8_+_128426535 | 0.01 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr7_-_150652924 | 0.01 |
ENST00000330883.4
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr5_-_32444828 | 0.01 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr11_-_68609377 | 0.01 |
ENST00000265641.5
ENST00000376618.2 |
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr5_-_65017921 | 0.01 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr18_+_43753974 | 0.01 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr8_+_54764346 | 0.01 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr4_+_41992489 | 0.01 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr12_+_69864129 | 0.01 |
ENST00000547219.1
ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2
|
fibroblast growth factor receptor substrate 2 |
| chr7_+_94285637 | 0.01 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr2_-_200322723 | 0.01 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr1_+_52521928 | 0.01 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr3_+_51575596 | 0.01 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr3_-_71774516 | 0.01 |
ENST00000425534.3
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr1_+_203274639 | 0.01 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr3_-_125094093 | 0.01 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr2_+_192542850 | 0.01 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr3_-_47823298 | 0.01 |
ENST00000254480.5
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr4_+_145567173 | 0.01 |
ENST00000296575.3
|
HHIP
|
hedgehog interacting protein |
| chr3_-_15901278 | 0.01 |
ENST00000399451.2
|
ANKRD28
|
ankyrin repeat domain 28 |
| chr10_+_101419187 | 0.01 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr1_-_85462623 | 0.01 |
ENST00000370608.3
|
MCOLN2
|
mucolipin 2 |
| chr7_-_151217001 | 0.01 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr17_+_35766965 | 0.01 |
ENST00000394395.2
ENST00000589153.1 ENST00000586023.1 |
TADA2A
|
transcriptional adaptor 2A |
| chr1_-_153588334 | 0.01 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr18_+_34409069 | 0.01 |
ENST00000543923.1
ENST00000280020.5 ENST00000435985.2 ENST00000592521.1 ENST00000587139.1 |
KIAA1328
|
KIAA1328 |
| chr1_+_29063271 | 0.01 |
ENST00000373812.3
|
YTHDF2
|
YTH domain family, member 2 |
| chr5_-_43313574 | 0.01 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chrX_-_70288234 | 0.01 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr2_+_155554797 | 0.01 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chrX_-_74376108 | 0.00 |
ENST00000339447.4
ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7
|
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr7_-_141401951 | 0.00 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr15_+_68346501 | 0.00 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr2_-_43453734 | 0.00 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr12_-_31479045 | 0.00 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr12_-_67072714 | 0.00 |
ENST00000545666.1
ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_+_202976493 | 0.00 |
ENST00000367242.3
|
TMEM183A
|
transmembrane protein 183A |
| chr12_+_27396901 | 0.00 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr1_+_161284047 | 0.00 |
ENST00000367975.2
ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr18_+_56338618 | 0.00 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr11_+_65101225 | 0.00 |
ENST00000528416.1
ENST00000415073.2 ENST00000252268.4 |
DPF2
|
D4, zinc and double PHD fingers family 2 |
| chr10_-_105615164 | 0.00 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr1_+_36273743 | 0.00 |
ENST00000373210.3
|
AGO4
|
argonaute RISC catalytic component 4 |
| chr3_-_79068594 | 0.00 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr22_+_32340481 | 0.00 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr1_-_35658736 | 0.00 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr10_+_22605304 | 0.00 |
ENST00000475460.2
ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1
COMMD3
|
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr7_+_138145076 | 0.00 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr5_+_74632993 | 0.00 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr9_+_131445928 | 0.00 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chrX_+_73641286 | 0.00 |
ENST00000587091.1
|
SLC16A2
|
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr11_+_48002076 | 0.00 |
ENST00000418331.2
ENST00000440289.2 |
PTPRJ
|
protein tyrosine phosphatase, receptor type, J |
| chr9_+_128024067 | 0.00 |
ENST00000461379.1
ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1
|
GTPase activating protein and VPS9 domains 1 |
| chr12_-_66524482 | 0.00 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr11_-_102962929 | 0.00 |
ENST00000260247.5
|
DCUN1D5
|
DCN1, defective in cullin neddylation 1, domain containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.0 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |