Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
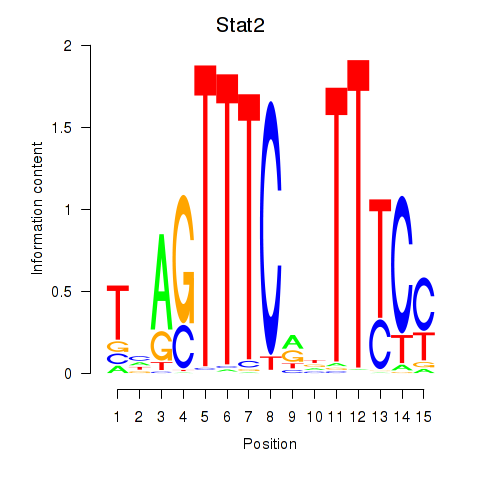
Results for Stat2
Z-value: 1.59
Transcription factors associated with Stat2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat2
|
ENSMUSG00000040033.17 | signal transducer and activator of transcription 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat2 | mm39_v1_chr10_+_128106414_128106446 | 0.80 | 6.1e-09 | Click! |
Activity profile of Stat2 motif
Sorted Z-values of Stat2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41498716 | 11.84 |
ENSMUST00000070380.5
|
Prss2
|
protease, serine 2 |
| chr19_+_3372296 | 9.04 |
ENSMUST00000237938.2
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr16_+_36041838 | 6.58 |
ENSMUST00000187183.7
ENSMUST00000187742.7 |
Csta2
|
cystatin A family member 2 |
| chr12_+_104372962 | 6.57 |
ENSMUST00000021506.6
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
| chr12_-_26506422 | 5.80 |
ENSMUST00000020970.10
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr18_+_60515755 | 5.70 |
ENSMUST00000237185.2
|
Iigp1
|
interferon inducible GTPase 1 |
| chr14_+_40827108 | 5.46 |
ENSMUST00000224514.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr19_+_34560922 | 5.37 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_-_48883053 | 5.29 |
ENSMUST00000059930.3
ENSMUST00000068063.4 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr6_+_121222865 | 5.19 |
ENSMUST00000032198.11
|
Usp18
|
ubiquitin specific peptidase 18 |
| chr11_-_83421333 | 5.05 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr14_+_78141679 | 4.35 |
ENSMUST00000022591.16
ENSMUST00000169978.2 ENSMUST00000227903.2 |
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr4_-_156285247 | 3.84 |
ENSMUST00000085425.6
|
Isg15
|
ISG15 ubiquitin-like modifier |
| chr2_-_173060647 | 3.82 |
ENSMUST00000109116.3
ENSMUST00000029018.14 |
Zbp1
|
Z-DNA binding protein 1 |
| chr4_+_34893772 | 3.80 |
ENSMUST00000029975.10
ENSMUST00000135871.8 ENSMUST00000108130.2 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr14_+_40826970 | 3.79 |
ENSMUST00000225720.2
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr11_+_58090382 | 3.79 |
ENSMUST00000035266.11
ENSMUST00000094169.11 ENSMUST00000168280.2 ENSMUST00000058704.9 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr1_+_85577709 | 3.77 |
ENSMUST00000155094.8
ENSMUST00000054279.15 ENSMUST00000147552.8 ENSMUST00000153574.8 ENSMUST00000150967.8 |
Sp100
|
nuclear antigen Sp100 |
| chr5_-_120915693 | 3.77 |
ENSMUST00000044833.9
|
Oas3
|
2'-5' oligoadenylate synthetase 3 |
| chr14_+_40827317 | 3.48 |
ENSMUST00000047286.7
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr1_-_173594475 | 3.26 |
ENSMUST00000111214.4
|
Ifi204
|
interferon activated gene 204 |
| chr12_+_26519203 | 3.11 |
ENSMUST00000020969.5
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr1_+_170846482 | 3.06 |
ENSMUST00000078825.5
|
Fcgr4
|
Fc receptor, IgG, low affinity IV |
| chr16_+_23428650 | 3.00 |
ENSMUST00000038423.6
ENSMUST00000211349.2 |
Rtp4
|
receptor transporter protein 4 |
| chr8_-_112064783 | 2.99 |
ENSMUST00000056157.14
ENSMUST00000120432.3 |
Mlkl
|
mixed lineage kinase domain-like |
| chrX_-_9335525 | 2.98 |
ENSMUST00000015484.10
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr7_+_142014546 | 2.90 |
ENSMUST00000105968.8
ENSMUST00000018963.11 ENSMUST00000105967.8 |
Lsp1
|
lymphocyte specific 1 |
| chr5_-_121045568 | 2.88 |
ENSMUST00000080322.8
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr16_-_35759461 | 2.86 |
ENSMUST00000081933.14
ENSMUST00000114885.3 |
Dtx3l
|
deltex 3-like, E3 ubiquitin ligase |
| chr19_+_34585322 | 2.85 |
ENSMUST00000076249.6
|
Ifit3b
|
interferon-induced protein with tetratricopeptide repeats 3B |
| chr18_+_60345152 | 2.75 |
ENSMUST00000031549.6
|
Gm4951
|
predicted gene 4951 |
| chr1_-_85526517 | 2.71 |
ENSMUST00000093508.7
|
Sp110
|
Sp110 nuclear body protein |
| chr11_-_118292678 | 2.54 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr12_+_37291728 | 2.54 |
ENSMUST00000160768.8
|
Agmo
|
alkylglycerol monooxygenase |
| chr15_+_79776597 | 2.54 |
ENSMUST00000175714.8
ENSMUST00000109620.10 ENSMUST00000165537.8 ENSMUST00000175752.8 ENSMUST00000176325.8 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr17_+_43700327 | 2.51 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chr13_+_74787952 | 2.50 |
ENSMUST00000221822.2
ENSMUST00000221526.2 |
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr5_-_92496730 | 2.49 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr1_-_173740467 | 2.35 |
ENSMUST00000009340.10
|
Ifi211
|
interferon activated gene 211 |
| chr4_-_46536088 | 2.35 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr6_+_145067457 | 2.34 |
ENSMUST00000032396.13
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr4_-_132125510 | 2.33 |
ENSMUST00000136711.2
ENSMUST00000084249.11 |
Phactr4
|
phosphatase and actin regulator 4 |
| chr1_+_85577766 | 2.30 |
ENSMUST00000066427.11
|
Sp100
|
nuclear antigen Sp100 |
| chr11_+_119283887 | 2.29 |
ENSMUST00000093902.12
ENSMUST00000131035.10 |
Rnf213
|
ring finger protein 213 |
| chr7_-_104114384 | 2.21 |
ENSMUST00000076922.6
|
Trim30a
|
tripartite motif-containing 30A |
| chr1_+_52158599 | 2.18 |
ENSMUST00000186574.7
ENSMUST00000070968.14 ENSMUST00000191435.7 ENSMUST00000186857.7 ENSMUST00000188681.7 |
Stat1
|
signal transducer and activator of transcription 1 |
| chr17_+_34406523 | 2.16 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr3_+_20011251 | 2.12 |
ENSMUST00000108328.8
|
Cp
|
ceruloplasmin |
| chr11_-_48762170 | 2.11 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr3_+_20011405 | 2.11 |
ENSMUST00000108325.9
|
Cp
|
ceruloplasmin |
| chr7_-_140846328 | 2.01 |
ENSMUST00000106023.8
ENSMUST00000097952.9 ENSMUST00000026571.11 |
Irf7
|
interferon regulatory factor 7 |
| chr1_-_173569301 | 2.00 |
ENSMUST00000042610.15
ENSMUST00000127730.2 |
Ifi207
|
interferon activated gene 207 |
| chr11_+_46701619 | 2.00 |
ENSMUST00000068877.7
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr11_-_70350783 | 2.00 |
ENSMUST00000019064.9
|
Cxcl16
|
chemokine (C-X-C motif) ligand 16 |
| chr4_-_40239700 | 2.00 |
ENSMUST00000142055.2
|
Ddx58
|
DEAD/H box helicase 58 |
| chr5_+_115034997 | 1.99 |
ENSMUST00000031542.13
ENSMUST00000146072.8 ENSMUST00000150361.2 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr1_-_173770010 | 1.95 |
ENSMUST00000042228.15
ENSMUST00000081216.12 ENSMUST00000129829.8 ENSMUST00000123708.8 |
Ifi203
|
interferon activated gene 203 |
| chr11_-_118292758 | 1.94 |
ENSMUST00000043722.10
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr19_+_12438125 | 1.93 |
ENSMUST00000081035.9
|
Mpeg1
|
macrophage expressed gene 1 |
| chr3_+_81839908 | 1.92 |
ENSMUST00000029649.3
|
Ctso
|
cathepsin O |
| chr16_+_35758836 | 1.88 |
ENSMUST00000114878.8
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr17_+_34406762 | 1.86 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr19_+_29344846 | 1.82 |
ENSMUST00000016640.8
|
Cd274
|
CD274 antigen |
| chr1_+_52158693 | 1.81 |
ENSMUST00000189347.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr18_-_33596890 | 1.80 |
ENSMUST00000237066.2
|
Nrep
|
neuronal regeneration related protein |
| chr10_-_94780695 | 1.79 |
ENSMUST00000099337.5
|
Plxnc1
|
plexin C1 |
| chr2_-_166904625 | 1.79 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr1_-_173707677 | 1.77 |
ENSMUST00000190651.4
ENSMUST00000188804.7 |
Mndal
|
myeloid nuclear differentiation antigen like |
| chr10_-_43880353 | 1.77 |
ENSMUST00000020017.14
|
Crybg1
|
crystallin beta-gamma domain containing 1 |
| chr12_-_79027531 | 1.75 |
ENSMUST00000174072.8
|
Tmem229b
|
transmembrane protein 229B |
| chr13_+_37529184 | 1.74 |
ENSMUST00000021860.7
|
Ly86
|
lymphocyte antigen 86 |
| chr4_+_135455427 | 1.73 |
ENSMUST00000102546.4
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chr6_+_128339882 | 1.73 |
ENSMUST00000073316.13
|
Foxm1
|
forkhead box M1 |
| chr17_-_35392254 | 1.71 |
ENSMUST00000025257.12
|
Aif1
|
allograft inflammatory factor 1 |
| chr3_+_20011201 | 1.71 |
ENSMUST00000091309.12
ENSMUST00000108329.8 ENSMUST00000003714.13 |
Cp
|
ceruloplasmin |
| chr12_+_108601963 | 1.71 |
ENSMUST00000223109.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr19_-_40502117 | 1.69 |
ENSMUST00000225148.2
ENSMUST00000224667.2 ENSMUST00000225153.2 ENSMUST00000238831.2 ENSMUST00000226047.2 ENSMUST00000224247.2 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr3_+_137377285 | 1.69 |
ENSMUST00000166899.3
|
Gm21962
|
predicted gene, 21962 |
| chr3_-_14843512 | 1.68 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr7_-_25488060 | 1.65 |
ENSMUST00000002677.11
ENSMUST00000085948.11 |
Axl
|
AXL receptor tyrosine kinase |
| chr2_+_155224105 | 1.63 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr4_+_94627755 | 1.61 |
ENSMUST00000071168.6
|
Tek
|
TEK receptor tyrosine kinase |
| chr18_+_61178211 | 1.60 |
ENSMUST00000025522.11
ENSMUST00000115274.2 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chr10_+_51367052 | 1.60 |
ENSMUST00000217705.2
ENSMUST00000078778.5 ENSMUST00000220182.2 ENSMUST00000220226.2 |
Lilrb4a
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4A |
| chr17_+_29879684 | 1.57 |
ENSMUST00000235014.2
ENSMUST00000130423.4 |
Cmtr1
|
cap methyltransferase 1 |
| chr1_-_173859321 | 1.54 |
ENSMUST00000059226.7
|
Ifi205
|
interferon activated gene 205 |
| chr12_-_79054050 | 1.54 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr2_-_180883813 | 1.53 |
ENSMUST00000094203.11
ENSMUST00000108831.8 |
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr19_-_43741363 | 1.51 |
ENSMUST00000045562.6
|
Cox15
|
cytochrome c oxidase assembly protein 15 |
| chr19_+_10819896 | 1.51 |
ENSMUST00000025646.3
|
Slc15a3
|
solute carrier family 15, member 3 |
| chr7_-_102214636 | 1.48 |
ENSMUST00000106913.3
ENSMUST00000033264.12 |
Trim21
|
tripartite motif-containing 21 |
| chr14_+_14475188 | 1.48 |
ENSMUST00000026315.8
|
Dnase1l3
|
deoxyribonuclease 1-like 3 |
| chr3_-_104725853 | 1.47 |
ENSMUST00000106775.8
ENSMUST00000166979.8 |
Mov10
|
Mov10 RISC complex RNA helicase |
| chr16_+_35759346 | 1.47 |
ENSMUST00000023622.13
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr3_+_89638044 | 1.45 |
ENSMUST00000029563.14
ENSMUST00000121094.8 ENSMUST00000118341.6 ENSMUST00000107405.6 |
Adar
|
adenosine deaminase, RNA-specific |
| chr14_+_11307729 | 1.42 |
ENSMUST00000160956.2
ENSMUST00000160340.8 ENSMUST00000162278.8 |
Fhit
|
fragile histidine triad gene |
| chr1_-_173318607 | 1.42 |
ENSMUST00000160565.4
|
Ifi206
|
interferon activated gene 206 |
| chr7_+_130179063 | 1.40 |
ENSMUST00000207918.2
ENSMUST00000215492.2 ENSMUST00000084513.12 ENSMUST00000059145.14 |
Tacc2
|
transforming, acidic coiled-coil containing protein 2 |
| chr7_-_104157006 | 1.40 |
ENSMUST00000033211.14
ENSMUST00000071069.13 |
Trim30d
|
tripartite motif-containing 30D |
| chr1_+_171216480 | 1.40 |
ENSMUST00000056449.9
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr10_+_128106414 | 1.40 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr3_-_104725581 | 1.39 |
ENSMUST00000168015.8
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr1_+_52158721 | 1.39 |
ENSMUST00000186057.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr15_+_81629258 | 1.39 |
ENSMUST00000109554.3
ENSMUST00000230946.2 |
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr11_+_72192455 | 1.37 |
ENSMUST00000151440.8
ENSMUST00000146233.8 ENSMUST00000140842.9 |
Xaf1
|
XIAP associated factor 1 |
| chr5_-_120887864 | 1.37 |
ENSMUST00000053909.13
ENSMUST00000081491.13 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr7_-_101667346 | 1.35 |
ENSMUST00000209844.2
ENSMUST00000211502.2 ENSMUST00000094134.5 |
Il18bp
|
interleukin 18 binding protein |
| chr7_+_100742070 | 1.35 |
ENSMUST00000208439.2
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr3_-_136031799 | 1.33 |
ENSMUST00000196159.5
ENSMUST00000041577.13 |
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr12_-_80307110 | 1.32 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr16_-_35691914 | 1.31 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr3_-_104725535 | 1.30 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr18_-_33596792 | 1.29 |
ENSMUST00000051087.16
|
Nrep
|
neuronal regeneration related protein |
| chr11_+_88890202 | 1.27 |
ENSMUST00000100627.9
ENSMUST00000107896.10 ENSMUST00000000284.7 |
Trim25
|
tripartite motif-containing 25 |
| chr4_+_94627513 | 1.26 |
ENSMUST00000073939.13
ENSMUST00000102798.8 |
Tek
|
TEK receptor tyrosine kinase |
| chr12_+_37292029 | 1.26 |
ENSMUST00000160390.2
|
Agmo
|
alkylglycerol monooxygenase |
| chr5_-_121025645 | 1.24 |
ENSMUST00000086368.12
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr7_+_78563184 | 1.24 |
ENSMUST00000121645.8
|
Isg20
|
interferon-stimulated protein |
| chr18_-_33597060 | 1.23 |
ENSMUST00000168890.2
|
Nrep
|
neuronal regeneration related protein |
| chr13_-_56326695 | 1.22 |
ENSMUST00000225063.2
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr10_+_122514669 | 1.21 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr9_-_58156982 | 1.20 |
ENSMUST00000135310.8
ENSMUST00000085673.11 ENSMUST00000114136.9 ENSMUST00000153820.8 ENSMUST00000124982.2 |
Pml
|
promyelocytic leukemia |
| chr19_+_6214416 | 1.19 |
ENSMUST00000045042.8
ENSMUST00000237511.2 |
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr16_+_26400454 | 1.19 |
ENSMUST00000096129.9
ENSMUST00000166294.9 ENSMUST00000174202.8 ENSMUST00000023156.13 |
Il1rap
|
interleukin 1 receptor accessory protein |
| chr3_+_142265787 | 1.19 |
ENSMUST00000199325.5
ENSMUST00000106222.9 |
Gbp3
|
guanylate binding protein 3 |
| chr12_+_108602008 | 1.19 |
ENSMUST00000172409.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr6_+_57557978 | 1.16 |
ENSMUST00000031817.10
|
Herc6
|
hect domain and RLD 6 |
| chr4_-_58911902 | 1.14 |
ENSMUST00000134848.2
ENSMUST00000107557.9 ENSMUST00000149301.8 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr4_+_138700195 | 1.11 |
ENSMUST00000123636.8
ENSMUST00000043042.10 ENSMUST00000050949.9 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr2_-_180883746 | 1.10 |
ENSMUST00000121484.2
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr11_-_48941949 | 1.05 |
ENSMUST00000132768.2
ENSMUST00000101295.9 ENSMUST00000093152.2 |
9930111J21Rik2
|
RIKEN cDNA 9930111J21 gene 2 |
| chr14_+_55842002 | 1.04 |
ENSMUST00000138037.2
|
Irf9
|
interferon regulatory factor 9 |
| chr11_+_72580823 | 1.04 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chr19_-_4384029 | 1.02 |
ENSMUST00000176653.2
|
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr9_-_58156935 | 1.02 |
ENSMUST00000124063.2
ENSMUST00000126690.8 |
Pml
|
promyelocytic leukemia |
| chr17_-_35081129 | 1.02 |
ENSMUST00000154526.8
|
Cfb
|
complement factor B |
| chr7_+_78563964 | 1.02 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr15_-_66684442 | 0.99 |
ENSMUST00000100572.10
|
Sla
|
src-like adaptor |
| chr2_-_35994072 | 0.99 |
ENSMUST00000112961.10
ENSMUST00000112966.10 |
Lhx6
|
LIM homeobox protein 6 |
| chr11_+_106642052 | 0.99 |
ENSMUST00000147326.9
ENSMUST00000182896.8 ENSMUST00000182908.8 ENSMUST00000086353.11 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr18_-_78185334 | 0.98 |
ENSMUST00000160639.2
|
Slc14a1
|
solute carrier family 14 (urea transporter), member 1 |
| chr4_-_40239778 | 0.98 |
ENSMUST00000037907.13
|
Ddx58
|
DEAD/H box helicase 58 |
| chr18_-_33596468 | 0.97 |
ENSMUST00000171533.9
|
Nrep
|
neuronal regeneration related protein |
| chr8_-_112064393 | 0.97 |
ENSMUST00000145862.3
|
Mlkl
|
mixed lineage kinase domain-like |
| chr12_+_37291632 | 0.97 |
ENSMUST00000049874.14
|
Agmo
|
alkylglycerol monooxygenase |
| chr4_-_136626073 | 0.97 |
ENSMUST00000046285.6
|
C1qa
|
complement component 1, q subcomponent, alpha polypeptide |
| chr14_+_48683581 | 0.96 |
ENSMUST00000227440.2
ENSMUST00000124720.8 ENSMUST00000226422.2 ENSMUST00000226400.2 |
Tmem260
|
transmembrane protein 260 |
| chr2_-_132095146 | 0.95 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr7_+_43057611 | 0.92 |
ENSMUST00000005592.7
|
Siglecg
|
sialic acid binding Ig-like lectin G |
| chr5_-_134258435 | 0.90 |
ENSMUST00000016094.13
ENSMUST00000111275.8 ENSMUST00000144086.2 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr16_-_43959361 | 0.89 |
ENSMUST00000124102.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr3_-_32670628 | 0.86 |
ENSMUST00000193050.2
ENSMUST00000108234.8 ENSMUST00000155737.8 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr13_-_56326511 | 0.84 |
ENSMUST00000169652.3
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr14_+_55815999 | 0.83 |
ENSMUST00000172738.2
ENSMUST00000089619.13 |
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_+_43365103 | 0.82 |
ENSMUST00000173002.8
ENSMUST00000057679.10 |
C1qtnf2
|
C1q and tumor necrosis factor related protein 2 |
| chr14_+_55815879 | 0.82 |
ENSMUST00000174563.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr14_+_55815580 | 0.80 |
ENSMUST00000174484.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr9_-_58065800 | 0.80 |
ENSMUST00000168864.4
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr5_+_34494272 | 0.78 |
ENSMUST00000182047.2
|
Rnf4
|
ring finger protein 4 |
| chr2_-_131953359 | 0.78 |
ENSMUST00000128899.2
|
Slc23a2
|
solute carrier family 23 (nucleobase transporters), member 2 |
| chr1_+_34511793 | 0.77 |
ENSMUST00000188972.3
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr17_+_34354556 | 0.77 |
ENSMUST00000236322.2
ENSMUST00000237402.2 |
H2-DMa
|
histocompatibility 2, class II, locus DMa |
| chr7_+_78563513 | 0.76 |
ENSMUST00000038142.15
|
Isg20
|
interferon-stimulated protein |
| chr11_-_48870105 | 0.76 |
ENSMUST00000141200.2
ENSMUST00000097494.9 ENSMUST00000093153.2 |
9930111J21Rik1
|
RIKEN cDNA 9930111J21 gene 1 |
| chr9_-_110819639 | 0.76 |
ENSMUST00000198702.2
|
Rtp3
|
receptor transporter protein 3 |
| chr18_+_65022035 | 0.75 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr3_+_94861386 | 0.75 |
ENSMUST00000107260.9
ENSMUST00000142311.8 ENSMUST00000137088.8 ENSMUST00000152869.8 ENSMUST00000107254.8 ENSMUST00000107253.8 |
Rfx5
|
regulatory factor X, 5 (influences HLA class II expression) |
| chr3_+_142266113 | 0.75 |
ENSMUST00000106221.8
|
Gbp3
|
guanylate binding protein 3 |
| chr1_+_173501215 | 0.73 |
ENSMUST00000085876.12
|
Ifi208
|
interferon activated gene 208 |
| chr3_-_59038031 | 0.73 |
ENSMUST00000091112.6
ENSMUST00000065220.13 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr17_+_37148015 | 0.73 |
ENSMUST00000179968.8
ENSMUST00000130367.8 ENSMUST00000053434.15 ENSMUST00000130801.8 ENSMUST00000144182.8 ENSMUST00000123715.8 |
Trim26
|
tripartite motif-containing 26 |
| chr11_+_115725140 | 0.72 |
ENSMUST00000173289.8
ENSMUST00000137900.2 |
Llgl2
|
LLGL2 scribble cell polarity complex component |
| chr5_-_90487583 | 0.72 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr9_+_107852733 | 0.71 |
ENSMUST00000035216.11
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr7_+_103893658 | 0.71 |
ENSMUST00000106849.9
ENSMUST00000060315.12 |
Trim34a
|
tripartite motif-containing 34A |
| chr17_-_35081456 | 0.70 |
ENSMUST00000025229.11
ENSMUST00000176203.9 ENSMUST00000128767.8 |
Cfb
|
complement factor B |
| chr16_-_43959993 | 0.70 |
ENSMUST00000137557.8
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr13_+_120761861 | 0.70 |
ENSMUST00000225029.2
|
BC147527
|
cDNA sequence BC147527 |
| chr2_-_152239966 | 0.70 |
ENSMUST00000063332.9
ENSMUST00000182625.2 |
Sox12
|
SRY (sex determining region Y)-box 12 |
| chr11_-_82882613 | 0.70 |
ENSMUST00000092840.11
ENSMUST00000038211.13 |
Slfn9
|
schlafen 9 |
| chr13_-_103901010 | 0.68 |
ENSMUST00000210489.2
|
Srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr14_-_63781381 | 0.67 |
ENSMUST00000058679.7
|
Mtmr9
|
myotubularin related protein 9 |
| chr7_+_75351713 | 0.67 |
ENSMUST00000207239.2
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr3_-_136031723 | 0.66 |
ENSMUST00000198206.2
|
Bank1
|
B cell scaffold protein with ankyrin repeats 1 |
| chr11_-_121120052 | 0.66 |
ENSMUST00000169393.8
ENSMUST00000106115.8 ENSMUST00000038709.14 ENSMUST00000147490.6 |
Cybc1
|
cytochrome b 245 chaperone 1 |
| chr3_+_142265904 | 0.65 |
ENSMUST00000128609.8
ENSMUST00000029935.14 |
Gbp3
|
guanylate binding protein 3 |
| chr7_+_78564062 | 0.64 |
ENSMUST00000205981.2
|
Isg20
|
interferon-stimulated protein |
| chr17_+_29879569 | 0.63 |
ENSMUST00000024816.13
ENSMUST00000235031.2 ENSMUST00000234911.2 |
Cmtr1
|
cap methyltransferase 1 |
| chr5_+_34493633 | 0.62 |
ENSMUST00000182709.8
ENSMUST00000030992.13 |
Rnf4
|
ring finger protein 4 |
| chr6_-_39095144 | 0.61 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chrX_+_80114242 | 0.59 |
ENSMUST00000171953.8
ENSMUST00000026760.3 |
Tmem47
|
transmembrane protein 47 |
| chr4_+_108691798 | 0.59 |
ENSMUST00000030296.9
|
Txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr2_-_120370333 | 0.59 |
ENSMUST00000171215.8
|
Zfp106
|
zinc finger protein 106 |
| chr14_+_55815817 | 0.56 |
ENSMUST00000174259.8
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr2_-_105229653 | 0.55 |
ENSMUST00000006128.7
|
Rcn1
|
reticulocalbin 1 |
| chr11_-_51748450 | 0.54 |
ENSMUST00000020655.14
ENSMUST00000109090.8 |
Jade2
|
jade family PHD finger 2 |
| chr17_-_34406193 | 0.52 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr17_+_34138699 | 0.52 |
ENSMUST00000234320.2
|
Tapbp
|
TAP binding protein |
| chr1_-_184464810 | 0.52 |
ENSMUST00000048572.7
|
Hlx
|
H2.0-like homeobox |
| chr4_-_42773987 | 0.50 |
ENSMUST00000095114.5
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0009087 | methionine catabolic process(GO:0009087) |
| 1.8 | 5.4 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
| 1.4 | 5.8 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 1.3 | 4.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 1.3 | 5.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.3 | 3.8 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 1.0 | 4.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 1.0 | 3.1 | GO:0006233 | dTDP biosynthetic process(GO:0006233) dTDP metabolic process(GO:0046072) |
| 0.6 | 2.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.6 | 3.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.6 | 3.0 | GO:0035546 | detection of virus(GO:0009597) interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.6 | 11.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.6 | 29.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.6 | 2.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.6 | 2.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.6 | 1.7 | GO:0014739 | positive regulation of muscle hyperplasia(GO:0014739) |
| 0.6 | 4.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.6 | 9.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.5 | 1.6 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.5 | 2.0 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.5 | 1.5 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.5 | 3.8 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.4 | 1.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.4 | 2.2 | GO:0097309 | cap1 mRNA methylation(GO:0097309) |
| 0.4 | 2.1 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.4 | 4.3 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.4 | 2.7 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.4 | 3.0 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.4 | 4.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.4 | 0.7 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.4 | 3.5 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.3 | 2.2 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.3 | 2.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.3 | 0.9 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.3 | 3.8 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 | 0.8 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.2 | 1.0 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.2 | 3.0 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.2 | 2.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 0.4 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.2 | 2.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.2 | 2.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.2 | 0.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.4 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.2 | 2.0 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.2 | 1.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 5.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.2 | 5.9 | GO:0046688 | copper ion transport(GO:0006825) response to copper ion(GO:0046688) |
| 0.2 | 1.4 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 1.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.7 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.1 | 0.4 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 1.5 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.1 | 2.5 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 1.0 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 6.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 3.1 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 1.7 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 1.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.0 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 2.0 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.1 | 1.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 0.7 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.5 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.4 | GO:0060337 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.4 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.1 | 0.4 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.7 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.7 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 2.2 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 1.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.1 | 0.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 2.4 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.1 | 1.2 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 1.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 0.7 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.1 | 0.8 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.1 | 2.5 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.1 | 2.4 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.1 | 0.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 1.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.1 | 2.5 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.2 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 1.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 1.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 4.2 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.4 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 1.7 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 3.0 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.2 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 1.0 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.5 | 4.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 3.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 1.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.3 | 1.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 2.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.3 | 1.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 9.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 3.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.7 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host cell(GO:0043657) |
| 0.1 | 1.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 2.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.6 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.0 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 10.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 5.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 12.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 6.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 12.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 3.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 3.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.1 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 17.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 3.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.7 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 3.0 | 9.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 1.8 | 5.3 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 1.7 | 5.1 | GO:0031726 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 1.6 | 4.8 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 1.3 | 4.0 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 1.0 | 3.1 | GO:0004798 | thymidylate kinase activity(GO:0004798) |
| 0.8 | 9.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.7 | 3.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 3.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.6 | 1.7 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.5 | 5.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 5.4 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 1.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.5 | 3.7 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.4 | 2.5 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 2.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 3.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 2.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.4 | 1.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.3 | 1.0 | GO:0032139 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.3 | 1.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.3 | 2.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.3 | 3.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 0.8 | GO:0008520 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.3 | 3.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 2.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 3.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.4 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.2 | 0.6 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.2 | 1.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 2.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 1.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 4.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 2.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 5.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.1 | 4.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 1.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 5.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 12.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 9.6 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 6.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 13.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 13.4 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 5.1 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 2.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 2.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 1.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.5 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.8 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 6.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 7.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 1.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 5.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 13.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 3.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 2.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 3.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.0 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.8 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.4 | 10.7 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.4 | 3.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.4 | 12.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 6.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 9.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 13.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 8.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.2 | 4.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 8.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 3.8 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 5.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 4.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 1.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 2.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 2.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 4.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.9 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 5.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.3 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 1.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 2.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |