Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
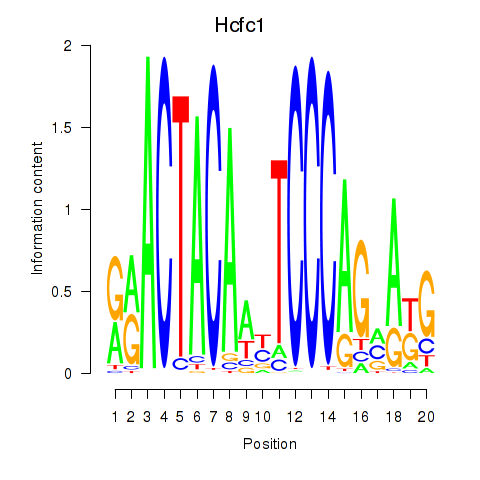
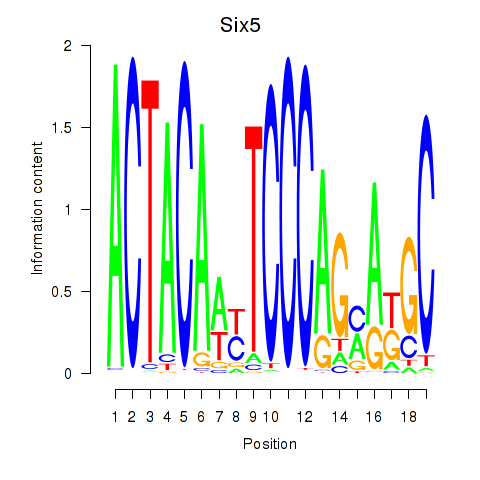
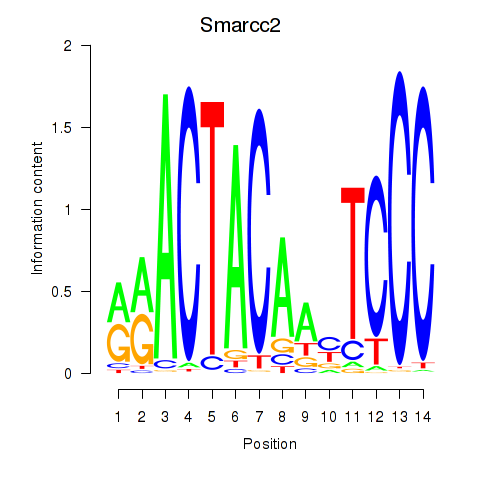
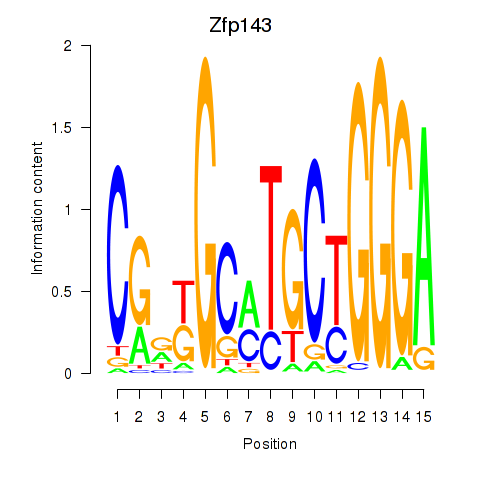
Results for Hcfc1_Six5_Smarcc2_Zfp143
Z-value: 3.00
Transcription factors associated with Hcfc1_Six5_Smarcc2_Zfp143
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hcfc1
|
ENSMUSG00000031386.15 | host cell factor C1 |
|
Six5
|
ENSMUSG00000040841.6 | sine oculis-related homeobox 5 |
|
Smarcc2
|
ENSMUSG00000025369.16 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
|
Zfp143
|
ENSMUSG00000061079.15 | zinc finger protein 143 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp143 | mm39_v1_chr7_+_109660887_109660950 | 0.79 | 9.4e-09 | Click! |
| Hcfc1 | mm39_v1_chrX_-_73009933_73009982 | 0.74 | 3.1e-07 | Click! |
| Smarcc2 | mm39_v1_chr10_+_128295159_128295197 | 0.60 | 1.0e-04 | Click! |
| Six5 | mm39_v1_chr7_+_18828519_18828558 | 0.18 | 2.8e-01 | Click! |
Activity profile of Hcfc1_Six5_Smarcc2_Zfp143 motif
Sorted Z-values of Hcfc1_Six5_Smarcc2_Zfp143 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Hcfc1_Six5_Smarcc2_Zfp143
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.4 | GO:0072355 | histone H3-T3 phosphorylation(GO:0072355) |
| 3.5 | 27.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 3.4 | 10.3 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 3.4 | 20.5 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 3.4 | 17.0 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 2.8 | 8.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.4 | 16.9 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 2.4 | 16.9 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 2.4 | 9.4 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 2.1 | 8.3 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 2.0 | 5.9 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 1.9 | 7.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 1.8 | 7.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 1.7 | 6.8 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 1.6 | 6.6 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 1.6 | 11.4 | GO:0051697 | protein delipidation(GO:0051697) |
| 1.6 | 4.8 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 1.5 | 10.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 1.4 | 12.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.3 | 5.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.3 | 9.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 1.3 | 3.9 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 1.3 | 3.8 | GO:0036245 | cellular response to menadione(GO:0036245) |
| 1.2 | 9.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 1.2 | 54.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 1.2 | 6.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 1.2 | 9.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 1.1 | 5.6 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 1.1 | 3.4 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 1.1 | 4.3 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.1 | 3.2 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 1.1 | 10.7 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 1.0 | 4.1 | GO:0017126 | nucleologenesis(GO:0017126) |
| 1.0 | 18.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 1.0 | 3.0 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 1.0 | 4.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 1.0 | 4.9 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.0 | 4.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.0 | 3.9 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.9 | 2.8 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.9 | 5.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.9 | 3.6 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) |
| 0.9 | 4.4 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.9 | 4.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.9 | 4.3 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.8 | 11.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.8 | 2.4 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.8 | 2.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.8 | 3.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.8 | 2.4 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.8 | 3.9 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.8 | 9.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.8 | 10.7 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.8 | 4.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.8 | 3.8 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.7 | 2.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.7 | 7.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.7 | 4.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.7 | 1.4 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.7 | 2.7 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.7 | 2.0 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.7 | 2.0 | GO:0032066 | nucleolus to nucleoplasm transport(GO:0032066) |
| 0.6 | 18.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.6 | 7.0 | GO:2000580 | regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.6 | 1.9 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.6 | 3.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.6 | 3.7 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.6 | 3.7 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.6 | 1.9 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.6 | 5.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.6 | 10.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.6 | 4.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.6 | 1.7 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.5 | 1.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.5 | 1.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.5 | 2.7 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.5 | 2.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.5 | 3.2 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.5 | 2.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.5 | 8.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.5 | 3.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.5 | 3.5 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.5 | 2.0 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.5 | 2.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.5 | 7.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.5 | 2.3 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.5 | 3.7 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.5 | 9.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.4 | 9.4 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.4 | 1.3 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.4 | 7.0 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.4 | 1.7 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.4 | 4.3 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.4 | 11.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.4 | 0.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.4 | 6.5 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.4 | 1.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.4 | 6.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.4 | 2.7 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.4 | 0.8 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.4 | 5.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.4 | 3.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.4 | 2.7 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.4 | 2.7 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.4 | 7.9 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.4 | 2.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.4 | 3.5 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.3 | 1.4 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 2.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 12.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.3 | 1.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.3 | 1.3 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.3 | 1.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.3 | 47.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.3 | 2.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.3 | 1.9 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.3 | 1.3 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.3 | 1.9 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.3 | 2.5 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.3 | 3.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 3.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.9 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.3 | 0.9 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 2.7 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.5 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.3 | 3.0 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.3 | 2.4 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.3 | 1.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.3 | 1.2 | GO:2001271 | regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.3 | 4.3 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.3 | 9.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.3 | 0.6 | GO:0009642 | response to light intensity(GO:0009642) response to high light intensity(GO:0009644) |
| 0.3 | 4.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 1.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.3 | 16.4 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.3 | 5.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) |
| 0.3 | 0.5 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 2.3 | GO:1904867 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.3 | 5.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.3 | 8.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 4.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 0.7 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 1.9 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.2 | 4.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 1.2 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 4.6 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.2 | 2.9 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 1.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 3.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) cap-dependent translational initiation(GO:0002191) |
| 0.2 | 14.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.2 | 4.9 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 0.6 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.2 | 0.6 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 3.7 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.2 | 2.8 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 1.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 0.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 0.2 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 | 1.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.6 | GO:0070839 | divalent metal ion export(GO:0070839) |
| 0.2 | 1.4 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 | 0.4 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.2 | 0.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.2 | 2.9 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 1.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.7 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.2 | 2.6 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 0.7 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.2 | 1.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.2 | 1.5 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.5 | GO:0030961 | peptidyl-arginine hydroxylation(GO:0030961) |
| 0.2 | 4.2 | GO:0048671 | negative regulation of collateral sprouting(GO:0048671) |
| 0.2 | 1.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 0.7 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 0.5 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.2 | 0.7 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 1.8 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 19.3 | GO:0007051 | spindle organization(GO:0007051) |
| 0.2 | 3.3 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.2 | 2.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 3.9 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.2 | 0.5 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) positive regulation of protein localization to centrosome(GO:1904781) |
| 0.2 | 2.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 | 5.5 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 0.8 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.2 | 10.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.2 | 4.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.2 | 2.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 1.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.2 | 4.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.1 | 2.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 2.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.3 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.1 | 1.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.3 | GO:1905216 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.1 | 2.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 2.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.5 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) |
| 0.1 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 1.6 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 | 1.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.8 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.3 | GO:0048822 | enucleate erythrocyte development(GO:0048822) |
| 0.1 | 0.5 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 | 0.9 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 15.2 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.1 | 0.3 | GO:0035037 | sperm entry(GO:0035037) |
| 0.1 | 1.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) |
| 0.1 | 4.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 2.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 41.8 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.1 | 2.3 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.1 | 1.7 | GO:0060352 | cell adhesion molecule production(GO:0060352) |
| 0.1 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.3 | GO:0061723 | glycophagy(GO:0061723) |
| 0.1 | 1.0 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 2.2 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.1 | 0.5 | GO:0018202 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.1 | 2.7 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 1.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 7.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 1.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 2.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 1.9 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.7 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 1.7 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.3 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 1.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 2.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 0.5 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.1 | 0.6 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.1 | 2.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 1.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.3 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0022417 | protein maturation by protein folding(GO:0022417) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 2.3 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 5.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.6 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.1 | 1.3 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.1 | 1.0 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.0 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 1.3 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.4 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.1 | 0.9 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 1.6 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 1.0 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.1 | 0.7 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 1.3 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 | 0.1 | GO:0014862 | regulation of the force of skeletal muscle contraction(GO:0014728) regulation of skeletal muscle contraction by chemo-mechanical energy conversion(GO:0014862) |
| 0.1 | 1.6 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.8 | GO:0039529 | RIG-I signaling pathway(GO:0039529) |
| 0.1 | 0.6 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 7.7 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.1 | 4.1 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 1.2 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.1 | 0.5 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.3 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 0.3 | GO:0061622 | glycolytic process through glucose-1-phosphate(GO:0061622) |
| 0.1 | 1.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.7 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 1.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.6 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.8 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 | 2.4 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.6 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.5 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 | 0.3 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.5 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.6 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.9 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 3.5 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.5 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.0 | 0.5 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 1.0 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.8 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.5 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.6 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.9 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 2.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:1903896 | regulation of IRE1-mediated unfolded protein response(GO:1903894) positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.4 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 1.7 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 1.2 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.7 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 1.3 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.5 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 2.9 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.6 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) rhombomere 4 development(GO:0021570) |
| 0.0 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.5 | GO:0070918 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.5 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.5 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 1.5 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.5 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.8 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 2.0 | GO:0050729 | positive regulation of inflammatory response(GO:0050729) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.8 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 3.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 6.6 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.4 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 1.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.6 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.0 | 0.5 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 1.1 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.3 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 1.0 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 2.0 | GO:0071375 | cellular response to peptide hormone stimulus(GO:0071375) |
| 0.0 | 0.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 16.9 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 2.8 | 8.3 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.4 | 9.8 | GO:1990298 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 2.0 | 6.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 2.0 | 7.9 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.8 | 23.6 | GO:0005818 | aster(GO:0005818) |
| 1.7 | 10.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 1.5 | 19.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 1.4 | 8.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.4 | 4.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.3 | 6.6 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 1.2 | 3.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 1.2 | 14.0 | GO:0000801 | central element(GO:0000801) |
| 1.0 | 9.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 1.0 | 4.8 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.9 | 8.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.9 | 10.1 | GO:0070578 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.9 | 5.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.8 | 3.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.8 | 4.8 | GO:0070449 | elongin complex(GO:0070449) |
| 0.8 | 5.6 | GO:0005638 | lamin filament(GO:0005638) |
| 0.8 | 7.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.7 | 5.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.7 | 5.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.7 | 6.5 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.7 | 2.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.7 | 10.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.7 | 2.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.6 | 3.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.6 | 3.6 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.6 | 5.4 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.6 | 4.6 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.6 | 3.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.6 | 27.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.5 | 4.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.5 | 3.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.5 | 3.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.5 | 7.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.5 | 2.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.5 | 5.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.5 | 1.4 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 3.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 11.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.4 | 5.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.4 | 4.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.4 | 4.3 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 4.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.4 | 4.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.4 | 8.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.4 | 1.8 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.4 | 1.4 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.4 | 7.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.4 | 6.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.3 | 11.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 9.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 1.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.3 | 11.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.3 | 2.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 6.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 25.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 3.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 14.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.3 | 5.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.3 | 6.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.3 | 3.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.3 | 3.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.3 | 3.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 22.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.3 | 16.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 0.9 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 2.0 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.3 | 2.8 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.3 | 1.4 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.3 | 24.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 1.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 0.8 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.3 | 3.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 1.3 | GO:0035363 | histone locus body(GO:0035363) |
| 0.3 | 10.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.3 | 2.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.3 | 3.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.2 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 2.4 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 5.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.2 | 2.8 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 3.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 3.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 3.9 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 2.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.7 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 2.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 4.8 | GO:0051286 | cell tip(GO:0051286) |
| 0.2 | 9.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.8 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 3.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 0.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 1.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.2 | 21.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 0.2 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.2 | 14.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 2.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.2 | 2.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 2.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 1.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 3.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.2 | 4.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.5 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 13.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.2 | 2.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 5.2 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.2 | 1.4 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.2 | 2.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 1.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 24.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 2.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.9 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 1.5 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 13.2 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 0.6 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 1.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 2.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 8.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 1.2 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.1 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 4.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 5.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 1.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.6 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 2.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 8.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 3.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 4.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.5 | GO:0048500 | signal recognition particle(GO:0048500) |
| 0.1 | 6.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 8.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 4.6 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 1.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 1.1 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.9 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 2.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 4.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.5 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 1.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 4.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 17.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 2.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 5.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 2.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 12.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 9.6 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 2.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 9.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 5.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 11.9 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.3 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.7 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.0 | 0.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 33.7 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 2.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.4 | GO:0035175 | histone kinase activity (H3-S10 specific)(GO:0035175) |
| 3.4 | 10.3 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 3.0 | 15.0 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 2.5 | 12.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 2.2 | 2.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 2.1 | 4.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 2.0 | 6.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 1.8 | 16.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 1.7 | 5.2 | GO:0003896 | DNA primase activity(GO:0003896) |
| 1.7 | 10.4 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 1.7 | 10.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 1.6 | 14.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 1.2 | 8.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 1.2 | 2.4 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 1.2 | 3.5 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 1.1 | 3.4 | GO:0005457 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 1.1 | 11.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 1.1 | 8.7 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 1.1 | 6.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 1.1 | 3.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.0 | 4.0 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 1.0 | 5.8 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.9 | 8.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.9 | 2.7 | GO:0030622 | U4atac snRNA binding(GO:0030622) |
| 0.9 | 3.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.9 | 2.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.9 | 7.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.9 | 2.6 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) |
| 0.9 | 8.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.8 | 4.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.8 | 37.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.8 | 8.0 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.8 | 10.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.8 | 5.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.7 | 2.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.7 | 3.6 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.7 | 4.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.7 | 2.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.7 | 1.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.6 | 4.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.6 | 1.9 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.6 | 8.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.6 | 1.9 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.6 | 4.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.6 | 4.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 3.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.6 | 7.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.5 | 3.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 1.1 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.5 | 2.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 3.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 5.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.5 | 2.0 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.5 | 10.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.5 | 1.9 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.5 | 51.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.4 | 0.9 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.4 | 2.6 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 4.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.4 | 7.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 9.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.4 | 4.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.4 | 4.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 2.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.4 | 10.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 4.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.3 | 2.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.3 | 2.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 3.8 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 17.4 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.3 | 1.7 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.3 | 2.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 6.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.3 | 1.0 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.3 | 1.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.3 | 3.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 7.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 1.3 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.3 | 3.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.3 | 8.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.3 | 1.5 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.3 | 6.0 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.3 | 11.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.3 | 2.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.3 | 1.5 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.3 | 6.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.3 | 0.8 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.3 | 2.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 6.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 21.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.2 | 10.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.2 | 26.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 1.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 6.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.3 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 1.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 2.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 5.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 5.5 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 5.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 1.8 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 1.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 1.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 1.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 4.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 5.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.2 | 0.6 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.2 | 2.9 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 2.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 8.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 4.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 2.4 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.2 | 1.4 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 0.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.2 | 0.8 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 0.6 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.2 | 0.8 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.2 | 2.8 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 3.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 3.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 9.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.6 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 3.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) phosphatidic acid binding(GO:0070300) |
| 0.1 | 1.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 3.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.9 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 2.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 21.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 4.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 10.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 3.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.3 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.1 | 6.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.7 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.7 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 5.5 | GO:0008186 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 2.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.8 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.6 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.1 | 3.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.9 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.1 | 2.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 3.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 3.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 3.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.8 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 1.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.7 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.1 | 4.7 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.1 | 0.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.6 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 1.8 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.3 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 7.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 1.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 2.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 27.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 3.4 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.4 | GO:0070990 | snRNP binding(GO:0070990) U1 snRNP binding(GO:1990446) |
| 0.0 | 1.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 2.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 3.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 10.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 15.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 2.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.1 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 17.3 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 5.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.4 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 67.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.8 | 38.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.5 | 26.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.5 | 18.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.4 | 18.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 9.4 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 6.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.3 | 8.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.3 | 4.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 5.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 4.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 9.6 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.2 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 3.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 8.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 5.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.1 | 2.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 12.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 8.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 6.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 10.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 5.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 4.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 2.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 3.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 4.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 5.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 3.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 33.6 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 1.1 | 30.5 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 1.0 | 2.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.9 | 20.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.8 | 10.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.7 | 16.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.7 | 9.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.7 | 3.5 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.7 | 6.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.7 | 7.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.6 | 69.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.6 | 13.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.5 | 14.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.4 | 11.8 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.4 | 8.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 58.4 | REACTOME 3 UTR MEDIATED TRANSLATIONAL REGULATION | Genes involved in 3' -UTR-mediated translational regulation |
| 0.4 | 0.8 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.4 | 11.2 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.4 | 7.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.3 | 9.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 5.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 7.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.3 | 8.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.3 | 20.7 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 3.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 3.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 7.8 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.2 | 6.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.2 | 6.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 3.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 6.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.2 | 4.5 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 4.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 0.2 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.2 | 2.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 4.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.6 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 2.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 2.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 4.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 2.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.9 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 8.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 2.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 8.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 3.2 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.5 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 1.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 8.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME SIGNALING BY NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 12.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.0 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.4 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.7 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |