Project
GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
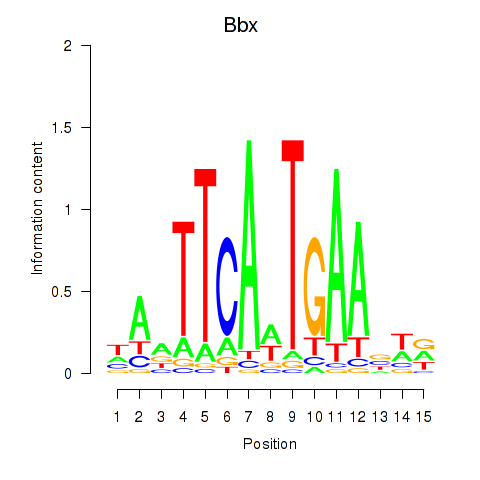
Results for Bbx
Z-value: 0.32
Transcription factors associated with Bbx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Bbx
|
ENSMUSG00000022641.16 | bobby sox HMG box containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bbx | mm39_v1_chr16_-_50252703_50252772 | 0.29 | 8.9e-02 | Click! |
Activity profile of Bbx motif
Sorted Z-values of Bbx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_41121979 | 4.20 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr12_+_103354915 | 2.45 |
ENSMUST00000101094.9
ENSMUST00000021620.13 |
Otub2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chrX_-_149597261 | 2.39 |
ENSMUST00000026302.13
ENSMUST00000129768.8 ENSMUST00000112699.9 |
Maged2
|
MAGE family member D2 |
| chr1_-_144651157 | 1.29 |
ENSMUST00000027603.4
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr2_-_152672535 | 1.14 |
ENSMUST00000146380.2
ENSMUST00000134902.2 ENSMUST00000134357.2 ENSMUST00000109820.5 |
Bcl2l1
|
BCL2-like 1 |
| chr2_-_152672185 | 1.03 |
ENSMUST00000140436.2
|
Bcl2l1
|
BCL2-like 1 |
| chr8_+_71069476 | 1.03 |
ENSMUST00000052437.6
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr10_+_85858050 | 0.89 |
ENSMUST00000120344.8
ENSMUST00000117597.2 |
Fbxo7
|
F-box protein 7 |
| chr10_+_85857726 | 0.76 |
ENSMUST00000130320.8
|
Fbxo7
|
F-box protein 7 |
| chr15_-_80929669 | 0.71 |
ENSMUST00000135047.2
|
Mrtfa
|
myocardin related transcription factor A |
| chr17_+_57665691 | 0.69 |
ENSMUST00000086763.13
ENSMUST00000233376.2 ENSMUST00000233840.2 ENSMUST00000232808.2 ENSMUST00000004850.8 |
Adgre1
|
adhesion G protein-coupled receptor E1 |
| chr13_-_52685305 | 0.60 |
ENSMUST00000057442.8
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2 |
| chrX_+_158491589 | 0.55 |
ENSMUST00000080394.13
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_-_97970388 | 0.44 |
ENSMUST00000120520.8
|
Acer3
|
alkaline ceramidase 3 |
| chr10_+_58207229 | 0.43 |
ENSMUST00000238939.2
|
Lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr15_-_11907267 | 0.33 |
ENSMUST00000228489.2
|
Npr3
|
natriuretic peptide receptor 3 |
| chr13_-_98152768 | 0.32 |
ENSMUST00000238746.2
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr15_-_50746202 | 0.29 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr13_-_91955781 | 0.27 |
ENSMUST00000022121.13
|
Zcchc9
|
zinc finger, CCHC domain containing 9 |
| chr12_-_86125793 | 0.27 |
ENSMUST00000003687.8
|
Tgfb3
|
transforming growth factor, beta 3 |
| chr13_+_46655324 | 0.26 |
ENSMUST00000021802.16
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_+_20511991 | 0.25 |
ENSMUST00000149543.9
ENSMUST00000232207.2 ENSMUST00000118919.9 |
Fam131a
|
family with sequence similarity 131, member A |
| chr6_+_67890534 | 0.19 |
ENSMUST00000197406.5
ENSMUST00000103311.3 |
Igkv11-125
|
immunoglobulin kappa variable 11-125 |
| chr9_-_38585897 | 0.18 |
ENSMUST00000215461.3
|
Olfr918
|
olfactory receptor 918 |
| chr11_-_79418500 | 0.15 |
ENSMUST00000154415.2
|
Evi2a
|
ecotropic viral integration site 2a |
| chr10_-_89270554 | 0.14 |
ENSMUST00000220071.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr18_-_60724855 | 0.12 |
ENSMUST00000056533.9
|
Myoz3
|
myozenin 3 |
| chr3_-_37286714 | 0.12 |
ENSMUST00000161015.2
ENSMUST00000029273.8 |
Il21
|
interleukin 21 |
| chr11_-_87788066 | 0.11 |
ENSMUST00000217095.2
ENSMUST00000215150.2 |
Olfr463
|
olfactory receptor 463 |
| chr13_-_65200204 | 0.11 |
ENSMUST00000222769.2
|
Prss47
|
protease, serine 47 |
| chr10_-_44334683 | 0.11 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr2_-_155434487 | 0.10 |
ENSMUST00000155347.2
ENSMUST00000130881.8 ENSMUST00000079691.13 |
Gss
|
glutathione synthetase |
| chr10_-_129741736 | 0.09 |
ENSMUST00000216182.2
|
Olfr815
|
olfactory receptor 815 |
| chr13_+_49761506 | 0.09 |
ENSMUST00000021822.7
|
Ogn
|
osteoglycin |
| chr15_+_30457772 | 0.07 |
ENSMUST00000228282.2
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2 |
| chr12_-_115944754 | 0.06 |
ENSMUST00000103551.2
|
Ighv1-84
|
immunoglobulin heavy variable 1-84 |
| chrX_+_67805443 | 0.06 |
ENSMUST00000069731.12
ENSMUST00000114647.8 |
Fmr1nb
|
Fmr1 neighbor |
| chr10_-_44334711 | 0.04 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr2_+_87185159 | 0.03 |
ENSMUST00000215163.3
|
Olfr1120
|
olfactory receptor 1120 |
| chrX_-_106859842 | 0.03 |
ENSMUST00000120722.2
|
2610002M06Rik
|
RIKEN cDNA 2610002M06 gene |
| chr14_+_53775648 | 0.03 |
ENSMUST00000200115.2
ENSMUST00000103650.3 |
Trav12-1
|
T cell receptor alpha variable 12-1 |
| chr10_-_89270527 | 0.02 |
ENSMUST00000218764.2
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr18_-_66103308 | 0.01 |
ENSMUST00000025397.7
|
Cplx4
|
complexin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Bbx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.5 | 2.2 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 2.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 2.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 1.7 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.1 | 0.3 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.4 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.4 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 2.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 2.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 4.2 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 1.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |