Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
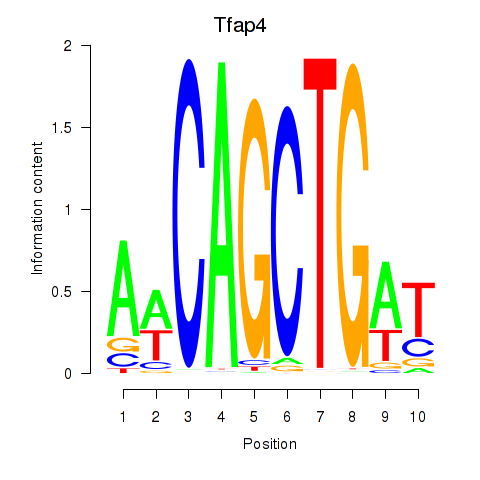
Results for Tfap4
Z-value: 1.59
Transcription factors associated with Tfap4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap4
|
ENSMUSG00000005718.9 | Tfap4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap4 | mm39_v1_chr16_-_4377640_4377718 | -0.64 | 3.0e-05 | Click! |
Activity profile of Tfap4 motif
Sorted Z-values of Tfap4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_97066937 | 13.26 |
ENSMUST00000043077.8
|
Thrsp
|
thyroid hormone responsive |
| chr3_+_14928561 | 8.36 |
ENSMUST00000029076.6
|
Car3
|
carbonic anhydrase 3 |
| chr7_+_107166653 | 6.23 |
ENSMUST00000120990.2
|
Olfml1
|
olfactomedin-like 1 |
| chr9_+_74883377 | 5.51 |
ENSMUST00000081746.7
|
Fam214a
|
family with sequence similarity 214, member A |
| chr6_-_124519240 | 5.13 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr6_-_72212547 | 4.75 |
ENSMUST00000042646.8
|
Atoh8
|
atonal bHLH transcription factor 8 |
| chr1_-_121255448 | 4.71 |
ENSMUST00000186915.2
ENSMUST00000160968.8 ENSMUST00000162582.2 |
Insig2
|
insulin induced gene 2 |
| chr2_+_155223728 | 4.65 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr1_-_121255400 | 4.45 |
ENSMUST00000159085.8
ENSMUST00000159125.2 ENSMUST00000161818.2 |
Insig2
|
insulin induced gene 2 |
| chr1_-_121255753 | 4.45 |
ENSMUST00000003818.14
|
Insig2
|
insulin induced gene 2 |
| chr11_-_4068779 | 4.34 |
ENSMUST00000003681.8
|
Sec14l2
|
SEC14-like lipid binding 2 |
| chr7_-_141015240 | 4.24 |
ENSMUST00000138865.8
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22 |
| chr12_-_83643883 | 3.98 |
ENSMUST00000221919.2
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr1_-_121255503 | 3.46 |
ENSMUST00000160688.2
|
Insig2
|
insulin induced gene 2 |
| chr12_-_83643964 | 3.18 |
ENSMUST00000048319.6
|
Zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr4_+_139038043 | 3.17 |
ENSMUST00000073787.7
|
Akr7a5
|
aldo-keto reductase family 7, member A5 (aflatoxin aldehyde reductase) |
| chr8_+_120121612 | 3.02 |
ENSMUST00000098367.5
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr18_+_60509101 | 2.89 |
ENSMUST00000032473.7
ENSMUST00000066912.13 |
Iigp1
|
interferon inducible GTPase 1 |
| chr12_-_28673259 | 2.67 |
ENSMUST00000220836.2
|
Colec11
|
collectin sub-family member 11 |
| chr17_+_43581220 | 2.66 |
ENSMUST00000047399.6
|
Adgrf1
|
adhesion G protein-coupled receptor F1 |
| chr4_+_11156411 | 2.63 |
ENSMUST00000029865.4
|
Trp53inp1
|
transformation related protein 53 inducible nuclear protein 1 |
| chr19_-_4548602 | 2.61 |
ENSMUST00000048482.8
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr10_+_75729237 | 2.45 |
ENSMUST00000009236.6
ENSMUST00000217811.2 |
Derl3
|
Der1-like domain family, member 3 |
| chr12_-_28673311 | 2.44 |
ENSMUST00000036136.9
|
Colec11
|
collectin sub-family member 11 |
| chr15_-_100497863 | 2.42 |
ENSMUST00000073837.13
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr2_+_155224105 | 2.41 |
ENSMUST00000134218.2
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr11_+_109376432 | 2.37 |
ENSMUST00000106697.8
|
Arsg
|
arylsulfatase G |
| chr7_-_119801327 | 2.36 |
ENSMUST00000033198.6
|
Crym
|
crystallin, mu |
| chr8_-_37200051 | 2.36 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr1_-_59276252 | 2.32 |
ENSMUST00000163058.2
ENSMUST00000160945.2 ENSMUST00000027178.13 |
Als2
|
alsin Rho guanine nucleotide exchange factor |
| chr9_+_108174052 | 2.31 |
ENSMUST00000035230.7
|
Amt
|
aminomethyltransferase |
| chr14_-_20231871 | 2.30 |
ENSMUST00000024011.10
|
Kcnk5
|
potassium channel, subfamily K, member 5 |
| chr15_+_25622611 | 2.22 |
ENSMUST00000110457.8
ENSMUST00000137601.8 |
Myo10
|
myosin X |
| chr13_+_64309675 | 2.20 |
ENSMUST00000021929.10
|
Habp4
|
hyaluronic acid binding protein 4 |
| chr9_-_78495250 | 2.13 |
ENSMUST00000117645.8
ENSMUST00000052441.12 |
Slc17a5
|
solute carrier family 17 (anion/sugar transporter), member 5 |
| chr8_+_105318067 | 2.10 |
ENSMUST00000059588.8
|
Pdp2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr10_+_60113449 | 2.08 |
ENSMUST00000105465.8
ENSMUST00000179238.8 ENSMUST00000177779.8 ENSMUST00000004316.15 |
Psap
|
prosaposin |
| chr8_+_3550450 | 2.02 |
ENSMUST00000004683.13
ENSMUST00000160338.2 |
Mcoln1
|
mucolipin 1 |
| chr8_+_75760301 | 1.99 |
ENSMUST00000165630.3
ENSMUST00000212651.2 ENSMUST00000212388.2 ENSMUST00000212299.2 ENSMUST00000078847.13 ENSMUST00000211869.2 |
Tom1
|
target of myb1 trafficking protein |
| chr4_-_19922599 | 1.98 |
ENSMUST00000029900.6
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr14_+_67470735 | 1.97 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr6_+_90442269 | 1.92 |
ENSMUST00000113530.4
|
Klf15
|
Kruppel-like factor 15 |
| chr11_-_3881960 | 1.89 |
ENSMUST00000109990.8
|
Tcn2
|
transcobalamin 2 |
| chr14_+_67470884 | 1.88 |
ENSMUST00000176161.8
|
Ebf2
|
early B cell factor 2 |
| chr2_-_77000878 | 1.86 |
ENSMUST00000111833.3
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr11_-_3881789 | 1.81 |
ENSMUST00000109992.8
ENSMUST00000109988.2 |
Tcn2
|
transcobalamin 2 |
| chr1_-_79739469 | 1.76 |
ENSMUST00000113512.8
ENSMUST00000113513.8 ENSMUST00000113515.8 ENSMUST00000113514.8 ENSMUST00000113510.8 ENSMUST00000113511.8 ENSMUST00000048820.14 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr9_-_24414423 | 1.72 |
ENSMUST00000142064.8
ENSMUST00000170356.2 |
Dpy19l1
|
dpy-19-like 1 (C. elegans) |
| chrX_+_139565657 | 1.70 |
ENSMUST00000112990.8
ENSMUST00000112988.8 |
Mid2
|
midline 2 |
| chr8_+_3550533 | 1.69 |
ENSMUST00000208306.2
|
Mcoln1
|
mucolipin 1 |
| chr12_-_86931529 | 1.64 |
ENSMUST00000038422.8
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr16_-_84970580 | 1.63 |
ENSMUST00000227737.2
ENSMUST00000226801.2 |
App
|
amyloid beta (A4) precursor protein |
| chr3_-_89230190 | 1.63 |
ENSMUST00000200436.2
ENSMUST00000029673.10 |
Efna3
|
ephrin A3 |
| chr2_-_174280811 | 1.62 |
ENSMUST00000016400.9
|
Ctsz
|
cathepsin Z |
| chr16_-_84970617 | 1.62 |
ENSMUST00000226232.2
ENSMUST00000227021.2 ENSMUST00000005406.12 ENSMUST00000227723.2 |
App
|
amyloid beta (A4) precursor protein |
| chr11_-_3881995 | 1.55 |
ENSMUST00000020710.11
ENSMUST00000109989.10 ENSMUST00000109991.8 ENSMUST00000109993.9 |
Tcn2
|
transcobalamin 2 |
| chr2_-_75768752 | 1.54 |
ENSMUST00000099996.5
|
Ttc30b
|
tetratricopeptide repeat domain 30B |
| chr7_-_127494750 | 1.53 |
ENSMUST00000033074.8
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr6_+_107506678 | 1.50 |
ENSMUST00000049285.10
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr7_+_30676465 | 1.50 |
ENSMUST00000058093.6
|
Fam187b
|
family with sequence similarity 187, member B |
| chr15_-_4025190 | 1.50 |
ENSMUST00000046633.10
|
AW549877
|
expressed sequence AW549877 |
| chr1_+_171052623 | 1.48 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr1_+_87254729 | 1.45 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr5_+_102872838 | 1.43 |
ENSMUST00000112853.8
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr9_-_63509699 | 1.41 |
ENSMUST00000171243.2
ENSMUST00000163982.8 ENSMUST00000163624.8 |
Iqch
|
IQ motif containing H |
| chr1_+_87133521 | 1.41 |
ENSMUST00000188796.7
ENSMUST00000027470.14 ENSMUST00000186038.2 |
Chrng
|
cholinergic receptor, nicotinic, gamma polypeptide |
| chr6_+_8259405 | 1.40 |
ENSMUST00000160705.8
ENSMUST00000159433.8 |
Umad1
|
UMAP1-MVP12 associated (UMA) domain containing 1 |
| chr12_+_73954678 | 1.39 |
ENSMUST00000110464.8
ENSMUST00000021530.8 |
Hif1a
|
hypoxia inducible factor 1, alpha subunit |
| chr11_-_113599778 | 1.37 |
ENSMUST00000106617.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr11_-_69576363 | 1.36 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr2_+_153334710 | 1.35 |
ENSMUST00000109783.2
|
4930404H24Rik
|
RIKEN cDNA 4930404H24 gene |
| chr3_+_135191366 | 1.35 |
ENSMUST00000029814.10
|
Manba
|
mannosidase, beta A, lysosomal |
| chr1_+_84817547 | 1.35 |
ENSMUST00000097672.4
|
Fbxo36
|
F-box protein 36 |
| chr16_+_48662894 | 1.34 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr19_+_12577317 | 1.28 |
ENSMUST00000181868.8
ENSMUST00000092931.7 |
Gm4952
|
predicted gene 4952 |
| chr8_+_84699580 | 1.27 |
ENSMUST00000005606.8
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr6_-_93890659 | 1.27 |
ENSMUST00000093769.8
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr19_+_34560922 | 1.25 |
ENSMUST00000102825.4
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr19_+_60800012 | 1.25 |
ENSMUST00000128357.8
ENSMUST00000119633.8 ENSMUST00000025957.9 |
Dennd10
|
DENN domain containing 10 |
| chr8_+_45960931 | 1.23 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr7_+_143838149 | 1.23 |
ENSMUST00000146006.3
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chrX_+_167819606 | 1.21 |
ENSMUST00000087016.11
ENSMUST00000112129.8 ENSMUST00000112131.9 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr7_-_44711075 | 1.17 |
ENSMUST00000007981.9
ENSMUST00000210500.2 ENSMUST00000210493.2 |
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr8_+_22550727 | 1.14 |
ENSMUST00000072572.13
ENSMUST00000110737.3 ENSMUST00000131624.2 |
Alg11
|
asparagine-linked glycosylation 11 (alpha-1,2-mannosyltransferase) |
| chr2_+_160573604 | 1.10 |
ENSMUST00000174885.2
ENSMUST00000109462.8 |
Plcg1
|
phospholipase C, gamma 1 |
| chr9_-_50605079 | 1.09 |
ENSMUST00000120622.2
|
Dixdc1
|
DIX domain containing 1 |
| chr16_+_14523696 | 1.08 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr3_-_84128160 | 1.08 |
ENSMUST00000154152.2
ENSMUST00000107693.9 ENSMUST00000107695.9 |
Trim2
|
tripartite motif-containing 2 |
| chr14_+_3575879 | 1.08 |
ENSMUST00000150727.8
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr16_+_43330630 | 1.07 |
ENSMUST00000114695.3
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_-_63509747 | 1.06 |
ENSMUST00000080527.12
ENSMUST00000042322.11 |
Iqch
|
IQ motif containing H |
| chr5_-_28415020 | 1.05 |
ENSMUST00000118882.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr14_+_54496683 | 1.04 |
ENSMUST00000041197.13
ENSMUST00000239403.2 ENSMUST00000197605.3 |
Abhd4
|
abhydrolase domain containing 4 |
| chr13_+_46571910 | 1.04 |
ENSMUST00000037923.5
|
Rbm24
|
RNA binding motif protein 24 |
| chr5_-_28415166 | 1.04 |
ENSMUST00000117098.2
|
Cnpy1
|
canopy FGF signaling regulator 1 |
| chr2_-_48839218 | 1.03 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr9_-_105009023 | 1.03 |
ENSMUST00000035179.9
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr18_-_39062201 | 1.03 |
ENSMUST00000134864.2
|
Fgf1
|
fibroblast growth factor 1 |
| chr4_+_155045372 | 1.03 |
ENSMUST00000049621.7
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr6_-_52185674 | 1.02 |
ENSMUST00000062829.9
|
Hoxa6
|
homeobox A6 |
| chr7_-_44711130 | 1.02 |
ENSMUST00000211337.2
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr18_+_38429792 | 1.01 |
ENSMUST00000237211.2
|
Rnf14
|
ring finger protein 14 |
| chr11_-_12362136 | 1.01 |
ENSMUST00000174874.8
|
Cobl
|
cordon-bleu WH2 repeat |
| chr18_+_60426444 | 1.01 |
ENSMUST00000171297.2
|
F830016B08Rik
|
RIKEN cDNA F830016B08 gene |
| chr5_-_103174794 | 1.00 |
ENSMUST00000128869.8
|
Mapk10
|
mitogen-activated protein kinase 10 |
| chr5_+_73071897 | 0.98 |
ENSMUST00000200785.2
|
Slain2
|
SLAIN motif family, member 2 |
| chr4_+_122910382 | 0.96 |
ENSMUST00000102649.4
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr5_+_107655487 | 0.96 |
ENSMUST00000143074.2
|
Gm42669
|
predicted gene 42669 |
| chr14_-_45456821 | 0.95 |
ENSMUST00000128484.8
ENSMUST00000147853.8 ENSMUST00000022377.11 ENSMUST00000143609.2 ENSMUST00000153383.8 ENSMUST00000139526.9 |
Txndc16
|
thioredoxin domain containing 16 |
| chr4_+_74170165 | 0.95 |
ENSMUST00000030102.12
|
Kdm4c
|
lysine (K)-specific demethylase 4C |
| chr9_+_121539395 | 0.95 |
ENSMUST00000035113.11
ENSMUST00000215966.2 ENSMUST00000215833.2 ENSMUST00000215104.2 |
Ss18l2
|
SS18, nBAF chromatin remodeling complex subunit like 2 |
| chr7_-_141456045 | 0.93 |
ENSMUST00000130439.3
|
Tollip
|
toll interacting protein |
| chr12_-_80483343 | 0.93 |
ENSMUST00000054145.8
|
Dcaf5
|
DDB1 and CUL4 associated factor 5 |
| chr7_+_143838191 | 0.89 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr6_-_115228800 | 0.89 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr6_-_93890520 | 0.88 |
ENSMUST00000203688.3
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr11_+_102727122 | 0.88 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr2_-_52225763 | 0.87 |
ENSMUST00000238288.2
ENSMUST00000238749.2 |
Neb
|
nebulin |
| chr5_-_147662798 | 0.87 |
ENSMUST00000110529.6
ENSMUST00000031653.12 |
Flt1
|
FMS-like tyrosine kinase 1 |
| chr7_+_27770655 | 0.85 |
ENSMUST00000138392.8
ENSMUST00000076648.8 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr2_+_48839505 | 0.85 |
ENSMUST00000112745.8
ENSMUST00000112754.8 |
Mbd5
|
methyl-CpG binding domain protein 5 |
| chrX_+_139565319 | 0.84 |
ENSMUST00000128809.2
|
Mid2
|
midline 2 |
| chr1_-_14380418 | 0.84 |
ENSMUST00000027066.13
ENSMUST00000168081.9 |
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr14_+_30601157 | 0.84 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr9_-_105008972 | 0.83 |
ENSMUST00000186925.2
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
| chr6_-_142647944 | 0.82 |
ENSMUST00000100827.5
ENSMUST00000087527.11 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr6_+_112436466 | 0.80 |
ENSMUST00000075477.8
|
Cav3
|
caveolin 3 |
| chr14_+_14091030 | 0.80 |
ENSMUST00000224529.2
|
Oit1
|
oncoprotein induced transcript 1 |
| chr3_+_9315662 | 0.79 |
ENSMUST00000155203.2
|
Zbtb10
|
zinc finger and BTB domain containing 10 |
| chr2_-_48839276 | 0.79 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr12_-_112905721 | 0.78 |
ENSMUST00000221497.2
ENSMUST00000002881.5 ENSMUST00000221397.2 |
Nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr9_-_75316625 | 0.78 |
ENSMUST00000168937.8
|
Mapk6
|
mitogen-activated protein kinase 6 |
| chr5_+_96357337 | 0.78 |
ENSMUST00000117766.8
|
Mrpl1
|
mitochondrial ribosomal protein L1 |
| chr14_+_14090981 | 0.77 |
ENSMUST00000022269.7
|
Oit1
|
oncoprotein induced transcript 1 |
| chr2_-_5068807 | 0.77 |
ENSMUST00000114996.8
|
Optn
|
optineurin |
| chr6_-_39095144 | 0.76 |
ENSMUST00000038398.7
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr11_-_49004584 | 0.76 |
ENSMUST00000203007.2
|
Olfr1396
|
olfactory receptor 1396 |
| chr10_+_128106414 | 0.75 |
ENSMUST00000085708.3
ENSMUST00000105238.10 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr14_+_45457168 | 0.75 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr17_+_14087827 | 0.75 |
ENSMUST00000239324.2
|
Afdn
|
afadin, adherens junction formation factor |
| chr1_-_14380327 | 0.75 |
ENSMUST00000080664.14
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr7_-_126046814 | 0.74 |
ENSMUST00000146973.2
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr7_-_141456092 | 0.73 |
ENSMUST00000055819.13
ENSMUST00000001950.12 |
Tollip
|
toll interacting protein |
| chr13_-_8921732 | 0.73 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr19_-_60456742 | 0.73 |
ENSMUST00000051277.4
|
Prlhr
|
prolactin releasing hormone receptor |
| chr15_-_75620060 | 0.72 |
ENSMUST00000062002.6
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr8_+_55003359 | 0.72 |
ENSMUST00000033918.4
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr13_-_116446166 | 0.72 |
ENSMUST00000036060.13
|
Isl1
|
ISL1 transcription factor, LIM/homeodomain |
| chr19_+_8816663 | 0.71 |
ENSMUST00000160556.8
|
Bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr6_+_55994473 | 0.71 |
ENSMUST00000052827.6
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr5_+_73071607 | 0.71 |
ENSMUST00000144843.8
|
Slain2
|
SLAIN motif family, member 2 |
| chr13_-_119545479 | 0.71 |
ENSMUST00000223268.2
|
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr11_-_76070325 | 0.70 |
ENSMUST00000167114.8
ENSMUST00000094015.11 ENSMUST00000108419.9 ENSMUST00000170730.3 ENSMUST00000129256.2 ENSMUST00000056601.11 |
Vps53
|
VPS53 GARP complex subunit |
| chr14_+_123897383 | 0.69 |
ENSMUST00000049681.14
|
Itgbl1
|
integrin, beta-like 1 |
| chr11_+_9068507 | 0.69 |
ENSMUST00000164791.8
ENSMUST00000130522.8 |
Upp1
|
uridine phosphorylase 1 |
| chr18_+_38429858 | 0.68 |
ENSMUST00000171461.3
ENSMUST00000235811.2 |
Rnf14
|
ring finger protein 14 |
| chr19_+_4560500 | 0.68 |
ENSMUST00000068004.13
ENSMUST00000224726.3 |
Pcx
|
pyruvate carboxylase |
| chr2_-_80277969 | 0.68 |
ENSMUST00000028389.4
|
Frzb
|
frizzled-related protein |
| chr6_-_93890237 | 0.67 |
ENSMUST00000204167.2
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr2_-_26962187 | 0.67 |
ENSMUST00000009358.9
|
Mymk
|
myomaker, myoblast fusion factor |
| chr14_+_3575406 | 0.67 |
ENSMUST00000124353.2
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr17_+_30223707 | 0.66 |
ENSMUST00000226208.3
|
Zfand3
|
zinc finger, AN1-type domain 3 |
| chr1_+_127234441 | 0.66 |
ENSMUST00000171405.2
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chr5_+_73071695 | 0.65 |
ENSMUST00000143829.5
|
Slain2
|
SLAIN motif family, member 2 |
| chr15_-_97665679 | 0.65 |
ENSMUST00000129223.9
ENSMUST00000126854.9 ENSMUST00000135080.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr2_-_160208977 | 0.65 |
ENSMUST00000099126.5
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) |
| chr18_+_38429688 | 0.65 |
ENSMUST00000170811.8
ENSMUST00000072376.13 ENSMUST00000237903.2 ENSMUST00000236116.2 ENSMUST00000237824.2 ENSMUST00000236982.2 ENSMUST00000235549.2 ENSMUST00000237416.2 ENSMUST00000237089.2 |
Rnf14
|
ring finger protein 14 |
| chr2_-_5068743 | 0.64 |
ENSMUST00000027986.5
|
Optn
|
optineurin |
| chr18_+_38430015 | 0.64 |
ENSMUST00000236319.2
|
Rnf14
|
ring finger protein 14 |
| chr14_-_36628263 | 0.64 |
ENSMUST00000183007.2
|
Ccser2
|
coiled-coil serine rich 2 |
| chr13_-_119545520 | 0.64 |
ENSMUST00000069902.13
ENSMUST00000099149.10 ENSMUST00000109204.8 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr15_-_97665524 | 0.63 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr1_-_16689527 | 0.63 |
ENSMUST00000182554.9
|
Ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr19_-_4447080 | 0.59 |
ENSMUST00000075856.11
ENSMUST00000176483.3 ENSMUST00000116571.9 |
Kdm2a
|
lysine (K)-specific demethylase 2A |
| chr4_-_49681954 | 0.59 |
ENSMUST00000029991.3
|
Ppp3r2
|
protein phosphatase 3, regulatory subunit B, alpha isoform (calcineurin B, type II) |
| chr18_+_53378806 | 0.58 |
ENSMUST00000025417.9
|
Snx24
|
sorting nexing 24 |
| chr2_-_77000936 | 0.58 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chrX_+_100492684 | 0.58 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr12_+_88689638 | 0.58 |
ENSMUST00000190626.7
ENSMUST00000167103.8 |
Nrxn3
|
neurexin III |
| chr7_+_30157704 | 0.57 |
ENSMUST00000126297.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr16_+_20408886 | 0.57 |
ENSMUST00000232279.2
ENSMUST00000232474.2 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr14_-_56809287 | 0.56 |
ENSMUST00000065302.8
|
Cenpj
|
centromere protein J |
| chr9_-_58444754 | 0.55 |
ENSMUST00000213722.2
|
Cd276
|
CD276 antigen |
| chr2_+_174602574 | 0.55 |
ENSMUST00000140908.2
|
Edn3
|
endothelin 3 |
| chrX_-_134594530 | 0.55 |
ENSMUST00000078605.7
|
Tcp11x2
|
t-complex 11 family, X-linked 2 |
| chr15_+_34238174 | 0.55 |
ENSMUST00000022867.5
ENSMUST00000226627.2 |
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr5_+_28047147 | 0.54 |
ENSMUST00000036227.7
|
Htr5a
|
5-hydroxytryptamine (serotonin) receptor 5A |
| chr2_+_97298002 | 0.54 |
ENSMUST00000059049.8
|
Lrrc4c
|
leucine rich repeat containing 4C |
| chr9_+_121606750 | 0.54 |
ENSMUST00000098272.4
|
Klhl40
|
kelch-like 40 |
| chr18_+_53378734 | 0.53 |
ENSMUST00000165032.9
|
Snx24
|
sorting nexing 24 |
| chr2_-_136958544 | 0.53 |
ENSMUST00000028735.8
|
Jag1
|
jagged 1 |
| chr8_+_13034245 | 0.53 |
ENSMUST00000110873.10
ENSMUST00000173006.8 ENSMUST00000145067.8 |
Mcf2l
|
mcf.2 transforming sequence-like |
| chr14_+_53756760 | 0.53 |
ENSMUST00000103585.4
|
Trav11
|
T cell receptor alpha variable 11 |
| chr3_-_88363704 | 0.53 |
ENSMUST00000141471.2
ENSMUST00000123753.8 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr7_-_23907518 | 0.53 |
ENSMUST00000086006.12
|
Zfp111
|
zinc finger protein 111 |
| chr6_+_21215472 | 0.52 |
ENSMUST00000081542.6
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2 |
| chr4_-_137137088 | 0.51 |
ENSMUST00000024200.7
|
Cela3a
|
chymotrypsin-like elastase family, member 3A |
| chr6_-_92458324 | 0.51 |
ENSMUST00000113445.2
|
Prickle2
|
prickle planar cell polarity protein 2 |
| chr1_-_16163506 | 0.51 |
ENSMUST00000145070.8
ENSMUST00000151004.2 |
4930444P10Rik
|
RIKEN cDNA 4930444P10 gene |
| chr18_+_65933586 | 0.50 |
ENSMUST00000025394.14
ENSMUST00000236847.2 ENSMUST00000153193.3 |
Sec11c
|
SEC11 homolog C, signal peptidase complex subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0071874 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 1.0 | 3.0 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.9 | 17.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.9 | 5.3 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.9 | 2.6 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.8 | 2.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 1.4 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 1.4 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.5 | 3.2 | GO:0044598 | polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.4 | 1.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.4 | 1.7 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 5.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.4 | 1.9 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.4 | 1.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.4 | 1.1 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.4 | 1.9 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.4 | 1.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 1.0 | GO:2000978 | auditory receptor cell fate determination(GO:0042668) negative regulation of forebrain neuron differentiation(GO:2000978) |
| 0.3 | 1.3 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.3 | 13.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.3 | 0.9 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.3 | 5.4 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.3 | 2.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.3 | 0.8 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 2.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.7 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 0.7 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 1.4 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.2 | 0.7 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 2.5 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.2 | 0.7 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.2 | 0.8 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.2 | 4.7 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.2 | 1.6 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.2 | 1.6 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.2 | 1.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 0.8 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.2 | 3.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 8.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.4 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 2.1 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.1 | 0.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 4.2 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.1 | 0.7 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 1.8 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 | 2.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.6 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 2.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 3.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 2.1 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 1.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.5 | GO:0007210 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) serotonin receptor signaling pathway(GO:0007210) |
| 0.1 | 0.9 | GO:0048597 | post-embryonic eye morphogenesis(GO:0048050) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.4 | GO:1900623 | positive regulation of mast cell cytokine production(GO:0032765) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0061073 | ciliary body morphogenesis(GO:0061073) Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 1.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 5.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.1 | 0.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.4 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) |
| 0.1 | 1.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.5 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.9 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.3 | GO:0090282 | positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.1 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 2.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 3.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.9 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 | 0.2 | GO:0099526 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 7.5 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.1 | 2.4 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 1.7 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 2.7 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.1 | GO:0090194 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) |
| 0.1 | 0.3 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 2.4 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.1 | 0.4 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 2.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 2.8 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.6 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 1.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.6 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 1.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.0 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 4.4 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.1 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.8 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.5 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.6 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.1 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 1.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 1.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 1.2 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.6 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.6 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.5 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.4 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 1.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 1.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.0 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 17.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 1.2 | 7.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.4 | 2.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 1.3 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.3 | 3.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.3 | 0.8 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 2.9 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 1.8 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 2.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 0.4 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.1 | 0.7 | GO:1990745 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.1 | 1.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 1.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.8 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.4 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 3.0 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 0.7 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 8.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.2 | GO:0044317 | rod spherule(GO:0044317) |
| 0.1 | 1.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 5.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.6 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 9.9 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 6.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.4 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 1.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.6 | 8.4 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.6 | 3.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.6 | 1.9 | GO:0035870 | dITP diphosphatase activity(GO:0035870) |
| 0.5 | 2.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 1.5 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.5 | 5.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.4 | 3.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 1.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.4 | 1.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 2.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.3 | 1.7 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.3 | 1.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.3 | 9.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.3 | 4.2 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.3 | 6.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 2.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 1.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.7 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 2.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.2 | 0.7 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 1.4 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 0.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.4 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 2.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 2.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 3.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 4.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 0.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.1 | 1.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.1 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 2.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.7 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.1 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.7 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 5.3 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.1 | 2.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 3.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 3.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 7.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 10.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 3.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 2.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.6 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.2 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 1.4 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 5.2 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 6.9 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 2.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 3.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 4.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 4.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.6 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.2 | 3.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 2.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.8 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 5.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.1 | 1.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.9 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 2.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.9 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.0 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 1.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 4.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.1 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |