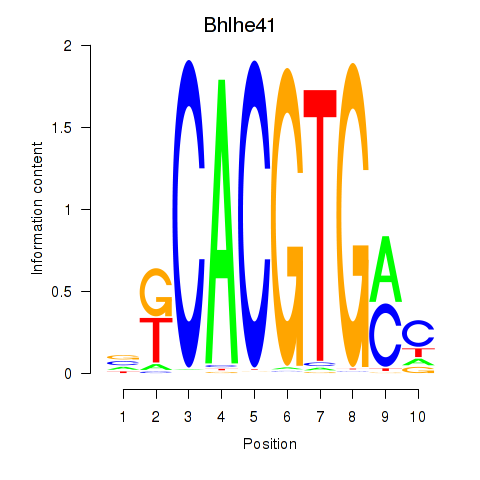
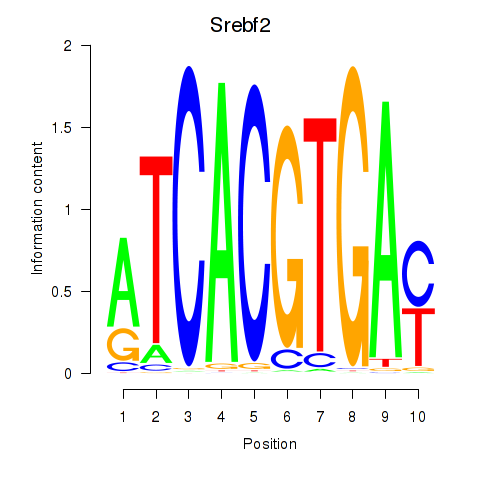
Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
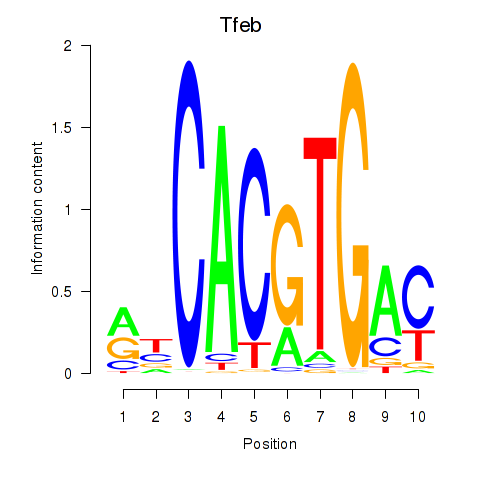
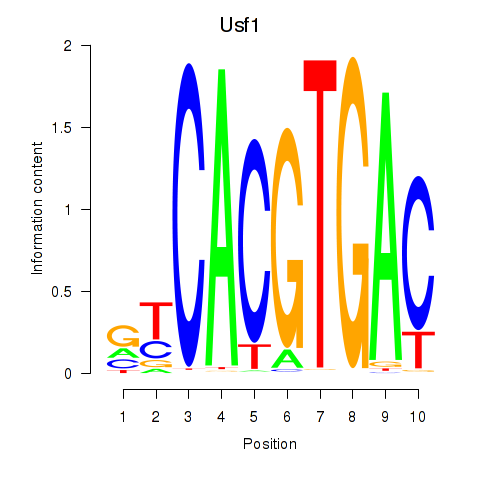
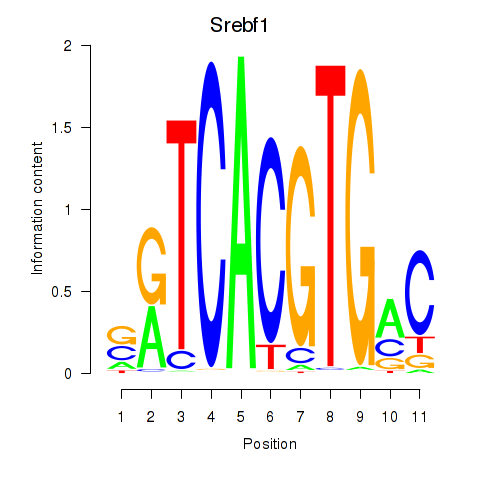
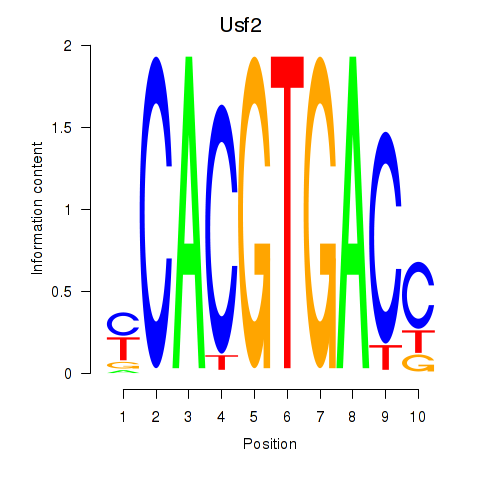
Results for Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Z-value: 1.71
Transcription factors associated with Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfeb
|
ENSMUSG00000023990.12 | transcription factor EB |
|
Usf1
|
ENSMUSG00000026641.7 | upstream transcription factor 1 |
|
Srebf1
|
ENSMUSG00000020538.9 | sterol regulatory element binding transcription factor 1 |
|
Usf2
|
ENSMUSG00000058239.7 | upstream transcription factor 2 |
|
Bhlhe41
|
ENSMUSG00000030256.5 | basic helix-loop-helix family, member e41 |
|
Srebf2
|
ENSMUSG00000022463.7 | sterol regulatory element binding factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srebf2 | mm10_v2_chr15_+_82147238_82147275 | 0.61 | 8.7e-05 | Click! |
| Srebf1 | mm10_v2_chr11_-_60220550_60220632 | -0.56 | 3.8e-04 | Click! |
| Usf2 | mm10_v2_chr7_-_30956742_30956803 | 0.52 | 1.0e-03 | Click! |
| Bhlhe41 | mm10_v2_chr6_-_145865483_145865558 | 0.30 | 7.3e-02 | Click! |
| Tfeb | mm10_v2_chr17_+_47737030_47737107 | 0.23 | 1.7e-01 | Click! |
| Usf1 | mm10_v2_chr1_+_171411343_171411384 | 0.19 | 2.7e-01 | Click! |
Activity profile of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 motif
Sorted Z-values of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_136068236 | 19.01 |
ENSMUST00000049130.7
|
Bex2
|
brain expressed X-linked 2 |
| chrX_-_136215443 | 13.94 |
ENSMUST00000113120.1
ENSMUST00000113118.1 ENSMUST00000058125.8 |
Bex1
|
brain expressed gene 1 |
| chr9_+_21368014 | 13.51 |
ENSMUST00000067646.4
ENSMUST00000115414.1 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chrX_+_136270302 | 10.96 |
ENSMUST00000113112.1
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chrX_+_136270253 | 10.78 |
ENSMUST00000178632.1
ENSMUST00000053540.4 |
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr11_+_116198853 | 10.16 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chrX_+_136138996 | 10.15 |
ENSMUST00000116527.1
|
Bex4
|
brain expressed gene 4 |
| chr2_+_30286383 | 9.66 |
ENSMUST00000064447.5
|
Nup188
|
nucleoporin 188 |
| chr13_+_108316395 | 9.54 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr13_+_108316332 | 9.45 |
ENSMUST00000051594.5
|
Depdc1b
|
DEP domain containing 1B |
| chr5_-_148995147 | 8.92 |
ENSMUST00000147473.1
|
Katnal1
|
katanin p60 subunit A-like 1 |
| chr12_-_32061221 | 8.74 |
ENSMUST00000003079.5
ENSMUST00000036497.9 |
Prkar2b
|
protein kinase, cAMP dependent regulatory, type II beta |
| chr9_-_22389113 | 8.28 |
ENSMUST00000040912.7
|
Anln
|
anillin, actin binding protein |
| chr7_+_90130227 | 8.20 |
ENSMUST00000049537.7
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr6_-_52217505 | 8.00 |
ENSMUST00000048715.6
|
Hoxa7
|
homeobox A7 |
| chr3_+_28781305 | 8.00 |
ENSMUST00000060500.7
|
Eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chrX_-_136203637 | 7.95 |
ENSMUST00000151592.1
ENSMUST00000131510.1 ENSMUST00000066819.4 |
Tceal5
|
transcription elongation factor A (SII)-like 5 |
| chr3_-_90052463 | 7.53 |
ENSMUST00000029553.9
ENSMUST00000064639.8 ENSMUST00000090908.6 |
Ubap2l
|
ubiquitin associated protein 2-like |
| chr19_-_10203880 | 7.35 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr6_+_128362919 | 7.29 |
ENSMUST00000073316.6
|
Foxm1
|
forkhead box M1 |
| chr3_+_159495408 | 7.27 |
ENSMUST00000120272.1
ENSMUST00000029825.7 ENSMUST00000106041.2 |
Depdc1a
|
DEP domain containing 1a |
| chr5_-_136170634 | 7.00 |
ENSMUST00000041048.1
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr1_-_193035651 | 6.95 |
ENSMUST00000016344.7
|
Syt14
|
synaptotagmin XIV |
| chr2_+_30286406 | 6.91 |
ENSMUST00000138666.1
ENSMUST00000113634.2 |
Nup188
|
nucleoporin 188 |
| chr4_-_134018829 | 6.86 |
ENSMUST00000051674.2
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_+_70000578 | 6.67 |
ENSMUST00000019362.8
|
Dvl2
|
dishevelled 2, dsh homolog (Drosophila) |
| chr5_+_118560719 | 6.60 |
ENSMUST00000100816.4
|
Med13l
|
mediator complex subunit 13-like |
| chr3_+_19644452 | 6.54 |
ENSMUST00000029139.7
|
Trim55
|
tripartite motif-containing 55 |
| chr8_+_75093591 | 6.54 |
ENSMUST00000005548.6
|
Hmox1
|
heme oxygenase (decycling) 1 |
| chr14_+_32321987 | 6.54 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chrX_+_153139941 | 6.53 |
ENSMUST00000039720.4
ENSMUST00000144175.2 |
Rragb
|
Ras-related GTP binding B |
| chr10_+_13090788 | 6.52 |
ENSMUST00000121646.1
ENSMUST00000121325.1 ENSMUST00000121766.1 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr1_-_75142360 | 6.32 |
ENSMUST00000041213.5
|
Cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr19_+_10018265 | 6.16 |
ENSMUST00000131407.1
|
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr17_-_12769605 | 6.06 |
ENSMUST00000024599.7
|
Igf2r
|
insulin-like growth factor 2 receptor |
| chr19_+_10018193 | 5.97 |
ENSMUST00000113161.2
ENSMUST00000117641.1 |
Rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chrX_+_136224035 | 5.95 |
ENSMUST00000113116.2
|
Tceal7
|
transcription elongation factor A (SII)-like 7 |
| chr17_-_66077022 | 5.91 |
ENSMUST00000150766.1
ENSMUST00000038116.5 |
Ankrd12
|
ankyrin repeat domain 12 |
| chr4_-_41275091 | 5.82 |
ENSMUST00000030143.6
ENSMUST00000108068.1 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr11_+_102881204 | 5.81 |
ENSMUST00000021307.3
ENSMUST00000159834.1 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr4_-_45084538 | 5.77 |
ENSMUST00000052236.6
|
Fbxo10
|
F-box protein 10 |
| chrX_-_135210672 | 5.73 |
ENSMUST00000033783.1
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr17_-_53689266 | 5.67 |
ENSMUST00000024736.7
|
Sgol1
|
shugoshin-like 1 (S. pombe) |
| chr15_-_96642883 | 5.59 |
ENSMUST00000088452.4
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr17_+_56040350 | 5.55 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr2_+_4300462 | 5.51 |
ENSMUST00000175669.1
|
Frmd4a
|
FERM domain containing 4A |
| chr11_-_94653964 | 5.48 |
ENSMUST00000039949.4
|
Eme1
|
essential meiotic endonuclease 1 homolog 1 (S. pombe) |
| chr17_-_26199008 | 5.38 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr2_+_84839395 | 5.16 |
ENSMUST00000146816.1
ENSMUST00000028469.7 |
Slc43a1
|
solute carrier family 43, member 1 |
| chr15_-_77956658 | 4.94 |
ENSMUST00000117725.1
ENSMUST00000016696.6 |
Foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr5_+_99979061 | 4.92 |
ENSMUST00000046721.1
|
4930524J08Rik
|
RIKEN cDNA 4930524J08 gene |
| chr19_-_4201591 | 4.89 |
ENSMUST00000025740.6
|
Rad9a
|
RAD9 homolog A |
| chrX_-_134751331 | 4.83 |
ENSMUST00000113194.1
ENSMUST00000052431.5 |
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chrX_-_134808984 | 4.82 |
ENSMUST00000035559.4
|
Armcx2
|
armadillo repeat containing, X-linked 2 |
| chr2_-_74578875 | 4.79 |
ENSMUST00000134168.1
ENSMUST00000111993.2 ENSMUST00000064503.6 |
Lnp
|
limb and neural patterns |
| chr7_+_16098458 | 4.77 |
ENSMUST00000006181.6
|
Napa
|
N-ethylmaleimide sensitive fusion protein attachment protein alpha |
| chr9_-_42124276 | 4.72 |
ENSMUST00000060989.8
|
Sorl1
|
sortilin-related receptor, LDLR class A repeats-containing |
| chr3_-_37724321 | 4.72 |
ENSMUST00000108105.1
ENSMUST00000079755.4 ENSMUST00000099128.1 |
Gm5148
|
predicted gene 5148 |
| chrX_+_134601179 | 4.68 |
ENSMUST00000074950.4
ENSMUST00000113203.1 ENSMUST00000113202.1 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chrX_+_136666375 | 4.64 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr15_+_59374198 | 4.63 |
ENSMUST00000079703.3
ENSMUST00000168722.1 |
Nsmce2
|
non-SMC element 2 homolog (MMS21, S. cerevisiae) |
| chr9_+_70679016 | 4.62 |
ENSMUST00000144537.1
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr6_+_116338013 | 4.61 |
ENSMUST00000079012.6
ENSMUST00000101032.3 |
March8
|
membrane-associated ring finger (C3HC4) 8 |
| chr11_+_95337012 | 4.61 |
ENSMUST00000037502.6
|
Fam117a
|
family with sequence similarity 117, member A |
| chr11_-_117780630 | 4.60 |
ENSMUST00000026659.3
ENSMUST00000127227.1 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr6_-_52217435 | 4.60 |
ENSMUST00000140316.1
|
Hoxa7
|
homeobox A7 |
| chrX_+_134601271 | 4.58 |
ENSMUST00000050331.6
ENSMUST00000059297.5 |
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2 |
| chrX_+_48146436 | 4.57 |
ENSMUST00000033427.6
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr7_-_46795661 | 4.51 |
ENSMUST00000123725.1
|
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr15_-_96642311 | 4.50 |
ENSMUST00000088454.5
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr15_+_73724754 | 4.49 |
ENSMUST00000163582.1
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr9_-_103480328 | 4.45 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chrX_+_56454871 | 4.43 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr4_+_148039035 | 4.36 |
ENSMUST00000097788.4
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase |
| chr6_-_88898664 | 4.32 |
ENSMUST00000058011.6
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae) |
| chr7_+_35802593 | 4.29 |
ENSMUST00000052454.2
|
E130304I02Rik
|
RIKEN cDNA E130304I02 gene |
| chr7_-_45466894 | 4.24 |
ENSMUST00000033093.8
|
Bax
|
BCL2-associated X protein |
| chr6_-_52226165 | 4.20 |
ENSMUST00000114425.2
|
Hoxa9
|
homeobox A9 |
| chr14_-_52197216 | 4.20 |
ENSMUST00000046709.7
|
Supt16
|
suppressor of Ty 16 |
| chr19_-_4615453 | 4.16 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr4_-_49597860 | 4.04 |
ENSMUST00000042750.2
|
Tmem246
|
transmembrane protein 246 |
| chr9_+_70678950 | 4.02 |
ENSMUST00000067880.6
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr2_-_74579379 | 4.01 |
ENSMUST00000130586.1
|
Lnp
|
limb and neural patterns |
| chr15_-_89425856 | 4.01 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr2_+_92915080 | 3.98 |
ENSMUST00000028648.2
|
Syt13
|
synaptotagmin XIII |
| chr5_+_143622440 | 3.94 |
ENSMUST00000116456.3
|
Cyth3
|
cytohesin 3 |
| chr11_+_61684419 | 3.94 |
ENSMUST00000093019.5
|
Fam83g
|
family with sequence similarity 83, member G |
| chr4_+_130308595 | 3.93 |
ENSMUST00000070532.7
|
Fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr16_-_92826004 | 3.91 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr5_-_124425572 | 3.87 |
ENSMUST00000168651.1
|
Sbno1
|
sno, strawberry notch homolog 1 (Drosophila) |
| chr6_-_128362812 | 3.85 |
ENSMUST00000112152.1
ENSMUST00000057421.8 ENSMUST00000112151.1 |
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chrX_+_50841434 | 3.81 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chr10_+_122678764 | 3.80 |
ENSMUST00000161487.1
ENSMUST00000067918.5 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr5_-_72168142 | 3.79 |
ENSMUST00000013693.6
|
Commd8
|
COMM domain containing 8 |
| chr4_-_49597425 | 3.78 |
ENSMUST00000150664.1
|
Tmem246
|
transmembrane protein 246 |
| chr2_-_160619971 | 3.76 |
ENSMUST00000109473.1
|
Gm14221
|
predicted gene 14221 |
| chr11_+_69913888 | 3.72 |
ENSMUST00000072581.2
ENSMUST00000116358.1 |
Gps2
|
G protein pathway suppressor 2 |
| chr4_+_148039097 | 3.71 |
ENSMUST00000141283.1
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase |
| chr3_+_137864573 | 3.70 |
ENSMUST00000174561.1
ENSMUST00000173790.1 |
H2afz
|
H2A histone family, member Z |
| chr5_-_113830422 | 3.70 |
ENSMUST00000100874.4
|
Selplg
|
selectin, platelet (p-selectin) ligand |
| chr19_-_7241216 | 3.69 |
ENSMUST00000025675.9
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr5_+_143622466 | 3.66 |
ENSMUST00000177196.1
|
Cyth3
|
cytohesin 3 |
| chrX_-_136172195 | 3.66 |
ENSMUST00000136533.1
ENSMUST00000146583.1 |
Tceal8
|
transcription elongation factor A (SII)-like 8 |
| chr2_+_129198757 | 3.64 |
ENSMUST00000028880.3
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr16_+_35770382 | 3.62 |
ENSMUST00000023555.4
|
Hspbap1
|
Hspb associated protein 1 |
| chrX_+_106027259 | 3.56 |
ENSMUST00000113557.1
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr9_-_103761820 | 3.56 |
ENSMUST00000049452.8
|
Tmem108
|
transmembrane protein 108 |
| chrX_+_106027300 | 3.52 |
ENSMUST00000055941.6
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr9_-_97111117 | 3.52 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr2_+_75659253 | 3.49 |
ENSMUST00000111964.1
ENSMUST00000111962.1 ENSMUST00000111961.1 ENSMUST00000164947.2 ENSMUST00000090792.4 |
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr4_-_45408646 | 3.47 |
ENSMUST00000153904.1
ENSMUST00000132815.2 ENSMUST00000107796.1 ENSMUST00000116341.3 |
Slc25a51
|
solute carrier family 25, member 51 |
| chr7_-_30821139 | 3.46 |
ENSMUST00000163504.1
|
Ffar2
|
free fatty acid receptor 2 |
| chrX_+_68678712 | 3.44 |
ENSMUST00000114654.1
ENSMUST00000114655.1 ENSMUST00000114657.2 ENSMUST00000114653.1 |
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr15_+_102296256 | 3.41 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr3_-_108722281 | 3.37 |
ENSMUST00000029482.9
|
Gpsm2
|
G-protein signalling modulator 2 (AGS3-like, C. elegans) |
| chr10_-_62792243 | 3.36 |
ENSMUST00000020268.5
|
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr11_+_69914179 | 3.33 |
ENSMUST00000057884.5
|
Gps2
|
G protein pathway suppressor 2 |
| chr19_-_4615647 | 3.32 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr17_-_33685386 | 3.31 |
ENSMUST00000139302.1
ENSMUST00000087582.5 ENSMUST00000114385.2 |
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr6_+_134929118 | 3.31 |
ENSMUST00000185152.1
ENSMUST00000184504.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chrX_+_136245065 | 3.21 |
ENSMUST00000048687.4
|
Wbp5
|
WW domain binding protein 5 |
| chr4_-_116994354 | 3.21 |
ENSMUST00000130273.1
|
Urod
|
uroporphyrinogen decarboxylase |
| chr17_+_48359891 | 3.20 |
ENSMUST00000024792.6
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr10_+_80016901 | 3.19 |
ENSMUST00000105373.1
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr6_+_134929089 | 3.19 |
ENSMUST00000183867.1
ENSMUST00000184991.1 ENSMUST00000183905.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr5_-_114773488 | 3.19 |
ENSMUST00000178440.1
ENSMUST00000043283.7 ENSMUST00000112185.2 |
Git2
|
G protein-coupled receptor kinase-interactor 2 |
| chr10_+_80016653 | 3.18 |
ENSMUST00000099501.3
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr15_+_79348061 | 3.12 |
ENSMUST00000163691.1
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr10_+_127063599 | 3.11 |
ENSMUST00000120226.1
ENSMUST00000133115.1 |
Cdk4
|
cyclin-dependent kinase 4 |
| chr9_-_13826946 | 3.10 |
ENSMUST00000147115.1
|
Cep57
|
centrosomal protein 57 |
| chr15_-_79834261 | 3.09 |
ENSMUST00000148358.1
|
Cbx6
|
chromobox 6 |
| chr4_-_148087961 | 3.08 |
ENSMUST00000030865.8
|
Agtrap
|
angiotensin II, type I receptor-associated protein |
| chr14_+_52197502 | 3.06 |
ENSMUST00000180857.1
|
Gm26590
|
predicted gene, 26590 |
| chr11_+_117782358 | 3.04 |
ENSMUST00000117781.1
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr11_+_117782076 | 2.99 |
ENSMUST00000127080.1
|
Tmc8
|
transmembrane channel-like gene family 8 |
| chr10_+_111164794 | 2.99 |
ENSMUST00000105275.1
ENSMUST00000095310.1 |
Osbpl8
|
oxysterol binding protein-like 8 |
| chr5_-_114773372 | 2.98 |
ENSMUST00000112183.1
ENSMUST00000086564.4 |
Git2
|
G protein-coupled receptor kinase-interactor 2 |
| chr8_+_85036906 | 2.95 |
ENSMUST00000093360.4
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr16_+_5050012 | 2.92 |
ENSMUST00000052449.5
|
Ubn1
|
ubinuclein 1 |
| chr6_+_72097561 | 2.90 |
ENSMUST00000069994.4
ENSMUST00000114112.1 |
St3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr3_+_137864487 | 2.90 |
ENSMUST00000041045.7
|
H2afz
|
H2A histone family, member Z |
| chr8_+_85037151 | 2.90 |
ENSMUST00000166592.1
|
Tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr11_+_117782281 | 2.89 |
ENSMUST00000050874.7
ENSMUST00000106334.2 |
Tmc8
|
transmembrane channel-like gene family 8 |
| chr10_-_120201558 | 2.87 |
ENSMUST00000020448.4
|
Irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr16_-_32487873 | 2.85 |
ENSMUST00000042042.7
|
Slc51a
|
solute carrier family 51, alpha subunit |
| chr5_-_99978914 | 2.85 |
ENSMUST00000112939.3
ENSMUST00000171786.1 ENSMUST00000072750.6 ENSMUST00000019128.8 ENSMUST00000172361.1 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr2_-_178414460 | 2.84 |
ENSMUST00000058678.4
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr2_+_91035613 | 2.84 |
ENSMUST00000111445.3
ENSMUST00000111446.3 ENSMUST00000050323.5 |
Rapsn
|
receptor-associated protein of the synapse |
| chr1_+_82724881 | 2.84 |
ENSMUST00000078332.6
|
Mff
|
mitochondrial fission factor |
| chr3_+_153844209 | 2.83 |
ENSMUST00000044089.3
|
Asb17
|
ankyrin repeat and SOCS box-containing 17 |
| chr4_-_116994374 | 2.82 |
ENSMUST00000030446.8
|
Urod
|
uroporphyrinogen decarboxylase |
| chr2_-_150668198 | 2.81 |
ENSMUST00000028944.3
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr14_+_70457447 | 2.80 |
ENSMUST00000003561.3
|
Phyhip
|
phytanoyl-CoA hydroxylase interacting protein |
| chr15_-_102510681 | 2.79 |
ENSMUST00000171565.1
|
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr11_-_102880925 | 2.78 |
ENSMUST00000021306.7
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr15_-_79834224 | 2.76 |
ENSMUST00000109623.1
ENSMUST00000109625.1 ENSMUST00000023060.6 ENSMUST00000089299.5 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr11_-_11462408 | 2.76 |
ENSMUST00000020413.3
|
Zpbp
|
zona pellucida binding protein |
| chr2_+_158794807 | 2.76 |
ENSMUST00000029186.7
ENSMUST00000109478.2 ENSMUST00000156893.1 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr19_-_8786272 | 2.76 |
ENSMUST00000176610.1
ENSMUST00000177056.1 |
Taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chrX_-_136172233 | 2.75 |
ENSMUST00000163584.1
ENSMUST00000060101.3 |
Tceal8
|
transcription elongation factor A (SII)-like 8 |
| chr10_+_119992916 | 2.75 |
ENSMUST00000105261.2
|
Grip1
|
glutamate receptor interacting protein 1 |
| chr11_+_87127267 | 2.74 |
ENSMUST00000139532.1
|
Trim37
|
tripartite motif-containing 37 |
| chr19_-_8786245 | 2.74 |
ENSMUST00000177216.1
|
Taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr19_+_34922351 | 2.73 |
ENSMUST00000087341.5
|
Kif20b
|
kinesin family member 20B |
| chr11_+_114727384 | 2.73 |
ENSMUST00000069325.7
|
Dnaic2
|
dynein, axonemal, intermediate chain 2 |
| chr10_+_86022189 | 2.72 |
ENSMUST00000120344.1
ENSMUST00000117597.1 |
Fbxo7
|
F-box protein 7 |
| chr8_+_119575235 | 2.71 |
ENSMUST00000093100.2
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr2_+_178414512 | 2.70 |
ENSMUST00000094251.4
|
Fam217b
|
family with sequence similarity 217, member B |
| chr1_+_17727034 | 2.70 |
ENSMUST00000159958.1
ENSMUST00000160305.1 ENSMUST00000095075.4 |
Crispld1
|
cysteine-rich secretory protein LCCL domain containing 1 |
| chr16_-_44139003 | 2.65 |
ENSMUST00000124102.1
|
Atp6v1a
|
ATPase, H+ transporting, lysosomal V1 subunit A |
| chr6_-_52158292 | 2.65 |
ENSMUST00000000964.5
ENSMUST00000120363.1 |
Hoxa1
|
homeobox A1 |
| chr17_-_56476462 | 2.61 |
ENSMUST00000067538.5
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S |
| chrX_+_68678541 | 2.60 |
ENSMUST00000088546.5
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr18_+_53176345 | 2.58 |
ENSMUST00000037850.5
|
Snx2
|
sorting nexin 2 |
| chr2_+_164074122 | 2.56 |
ENSMUST00000018353.7
|
Stk4
|
serine/threonine kinase 4 |
| chr14_-_67008834 | 2.55 |
ENSMUST00000111115.1
ENSMUST00000022634.8 |
Bnip3l
|
BCL2/adenovirus E1B interacting protein 3-like |
| chr4_+_130055010 | 2.54 |
ENSMUST00000123617.1
|
Col16a1
|
collagen, type XVI, alpha 1 |
| chr3_+_116562965 | 2.54 |
ENSMUST00000029573.5
|
Lrrc39
|
leucine rich repeat containing 39 |
| chrX_+_134686519 | 2.52 |
ENSMUST00000124226.2
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr11_-_102880981 | 2.52 |
ENSMUST00000107060.1
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr10_+_86021961 | 2.49 |
ENSMUST00000130320.1
|
Fbxo7
|
F-box protein 7 |
| chrX_+_8271381 | 2.48 |
ENSMUST00000033512.4
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr14_+_80000292 | 2.47 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr19_-_8786408 | 2.46 |
ENSMUST00000176496.1
|
Taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr19_-_60790692 | 2.46 |
ENSMUST00000025955.6
|
Eif3a
|
eukaryotic translation initiation factor 3, subunit A |
| chr1_-_40085823 | 2.46 |
ENSMUST00000181756.1
|
Gm16894
|
predicted gene, 16894 |
| chr11_-_34783850 | 2.45 |
ENSMUST00000093193.5
ENSMUST00000101365.2 |
Dock2
|
dedicator of cyto-kinesis 2 |
| chr11_-_96943945 | 2.45 |
ENSMUST00000107629.1
ENSMUST00000018803.5 |
Pnpo
|
pyridoxine 5'-phosphate oxidase |
| chr12_-_101958148 | 2.45 |
ENSMUST00000159883.1
ENSMUST00000160251.1 ENSMUST00000161011.1 ENSMUST00000021606.5 |
Atxn3
|
ataxin 3 |
| chrX_+_8271133 | 2.45 |
ENSMUST00000127103.1
ENSMUST00000115591.1 |
Slc38a5
|
solute carrier family 38, member 5 |
| chr18_-_12941777 | 2.45 |
ENSMUST00000122175.1
|
Osbpl1a
|
oxysterol binding protein-like 1A |
| chr2_+_84980458 | 2.43 |
ENSMUST00000028467.5
|
Prg2
|
proteoglycan 2, bone marrow |
| chr16_-_18811615 | 2.42 |
ENSMUST00000096990.3
|
Cdc45
|
cell division cycle 45 |
| chr4_-_48473403 | 2.42 |
ENSMUST00000164866.1
ENSMUST00000030030.8 |
Tex10
|
testis expressed gene 10 |
| chr4_-_141053660 | 2.42 |
ENSMUST00000040222.7
|
Crocc
|
ciliary rootlet coiled-coil, rootletin |
| chr14_-_104522615 | 2.41 |
ENSMUST00000022716.2
|
Rnf219
|
ring finger protein 219 |
| chr3_-_153725062 | 2.38 |
ENSMUST00000064460.5
|
St6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr1_+_82724909 | 2.38 |
ENSMUST00000073025.5
ENSMUST00000161648.1 ENSMUST00000160786.1 ENSMUST00000162003.1 |
Mff
|
mitochondrial fission factor |
| chr5_+_104046980 | 2.37 |
ENSMUST00000134313.1
|
Nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr1_+_183297060 | 2.37 |
ENSMUST00000109166.2
|
Aida
|
axin interactor, dorsalization associated |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfeb_Usf1_Srebf1_Usf2_Bhlhe41_Srebf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 24.8 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 2.7 | 8.1 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 2.6 | 12.9 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 2.4 | 7.1 | GO:0051542 | elastin biosynthetic process(GO:0051542) regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.2 | 6.5 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 2.0 | 8.1 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 1.9 | 5.8 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 1.7 | 8.6 | GO:0042117 | monocyte activation(GO:0042117) |
| 1.7 | 12.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 1.7 | 10.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 1.4 | 4.2 | GO:0002352 | B cell negative selection(GO:0002352) B cell homeostatic proliferation(GO:0002358) |
| 1.4 | 11.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 1.3 | 6.7 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 1.3 | 3.9 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 1.2 | 3.5 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 1.1 | 6.9 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 1.1 | 4.5 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 1.1 | 5.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.1 | 12.6 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 1.0 | 4.1 | GO:0007066 | female meiosis sister chromatid cohesion(GO:0007066) |
| 1.0 | 6.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 1.0 | 3.0 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 1.0 | 3.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 1.0 | 4.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 1.0 | 8.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.9 | 2.8 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.9 | 2.7 | GO:1903436 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.9 | 6.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.9 | 2.6 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.9 | 4.3 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.8 | 3.4 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.8 | 4.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.8 | 2.5 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.8 | 3.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.8 | 8.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.8 | 2.4 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.8 | 6.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.8 | 2.4 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.8 | 2.4 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.7 | 2.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.7 | 8.2 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.7 | 1.5 | GO:0033864 | positive regulation of NAD(P)H oxidase activity(GO:0033864) |
| 0.7 | 2.1 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.7 | 3.4 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.7 | 2.0 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.7 | 4.7 | GO:0030421 | defecation(GO:0030421) |
| 0.6 | 2.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.6 | 2.6 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.6 | 2.4 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.6 | 6.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 5.2 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.6 | 5.2 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.6 | 1.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.6 | 1.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.5 | 2.7 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.5 | 1.6 | GO:0046436 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.5 | 5.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.5 | 2.6 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.5 | 0.5 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.5 | 8.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 1.5 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.5 | 4.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.5 | 1.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.5 | 1.4 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.5 | 2.8 | GO:0051790 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.5 | 10.8 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.5 | 5.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 1.8 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.5 | 3.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 0.9 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.4 | 1.8 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.4 | 1.8 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.4 | 2.2 | GO:1902606 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.4 | 1.3 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 3.4 | GO:0002840 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) |
| 0.4 | 0.8 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.4 | 1.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 2.5 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.4 | 2.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.4 | 4.8 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.4 | 3.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.4 | 1.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 3.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.4 | 4.6 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.4 | 1.5 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.4 | 13.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.4 | 1.9 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.4 | 1.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.4 | 5.6 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.4 | 3.0 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.4 | 0.4 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.4 | 1.8 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.4 | 2.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.4 | 2.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.3 | 0.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 2.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 1.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 2.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 2.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 0.6 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.3 | 1.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.3 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 0.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 6.9 | GO:0032460 | negative regulation of protein oligomerization(GO:0032460) |
| 0.3 | 0.9 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.3 | 3.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.3 | 0.9 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.3 | 1.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.3 | 3.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.3 | 4.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.3 | 2.6 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.3 | 4.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 0.6 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.3 | 3.4 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.3 | 0.8 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 13.4 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.3 | 0.8 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.3 | 0.6 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.3 | 1.4 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.3 | 1.4 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.3 | 6.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.3 | 10.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.3 | 1.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.3 | 4.8 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.3 | 8.8 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.3 | 0.5 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.3 | 4.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.3 | 5.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.3 | 1.5 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.3 | 1.8 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.3 | 2.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 0.8 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 1.0 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 1.5 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 4.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 1.7 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.2 | 2.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 2.1 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 2.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.7 | GO:0002436 | immune complex clearance by monocytes and macrophages(GO:0002436) astrocyte chemotaxis(GO:0035700) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) negative regulation of eosinophil activation(GO:1902567) positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.2 | 0.5 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 5.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 10.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.2 | 2.5 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.2 | 3.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.4 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 1.5 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 | 2.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.2 | 0.4 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.2 | 1.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.2 | 0.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.2 | 0.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 0.6 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 1.0 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.2 | 1.4 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.2 | 1.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 0.6 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 3.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 1.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 1.1 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 2.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 6.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 2.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 0.2 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.2 | 0.7 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 | 3.9 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.2 | 0.5 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.2 | 1.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.2 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 11.1 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 3.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.1 | 0.3 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.1 | 1.0 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.6 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.1 | 4.1 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.1 | 0.8 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.4 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.6 | GO:0021586 | pons maturation(GO:0021586) |
| 0.1 | 1.4 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.1 | 4.3 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.3 | GO:0060003 | copper ion export(GO:0060003) |
| 0.1 | 1.6 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0070340 | detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.5 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.6 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.1 | 1.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 16.8 | GO:0030177 | positive regulation of Wnt signaling pathway(GO:0030177) |
| 0.1 | 3.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 3.4 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.7 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 2.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 11.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 5.8 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 2.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.4 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.5 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 | 0.9 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.6 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.7 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 1.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.7 | GO:0071971 | extracellular exosome assembly(GO:0071971) regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 0.8 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0071503 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 | 0.4 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.8 | GO:2000809 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 1.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.8 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 0.2 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 1.0 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.6 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.4 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 5.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.1 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.1 | 0.7 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 4.1 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.5 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.2 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 | 0.1 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.5 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.1 | 5.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 1.0 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 3.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 0.2 | GO:1904579 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.1 | 2.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 4.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 1.4 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.1 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.5 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.1 | 2.1 | GO:0048255 | mRNA stabilization(GO:0048255) |
| 0.1 | 2.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 1.7 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.5 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.1 | 0.7 | GO:1901978 | peripheral nervous system myelin maintenance(GO:0032287) positive regulation of spindle checkpoint(GO:0090232) positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 | 1.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 2.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.1 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 | 0.9 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.6 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 3.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 1.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 0.8 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.0 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 1.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 12.9 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 0.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 2.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 1.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.6 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 1.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 3.6 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 0.4 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 3.1 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 0.4 | GO:0014857 | skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 3.0 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 0.2 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.1 | 0.3 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 0.2 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 1.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.7 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.4 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0046005 | regulation of circadian sleep/wake cycle, wakefulness(GO:0010840) positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) circadian sleep/wake cycle, wakefulness(GO:0042746) positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.7 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 1.2 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.6 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.0 | 0.3 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.9 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 3.5 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.4 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 2.8 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0050691 | regulation of defense response to virus by host(GO:0050691) |
| 0.0 | 0.1 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.7 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 1.3 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.4 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0071868 | cellular response to monoamine stimulus(GO:0071868) cellular response to catecholamine stimulus(GO:0071870) |
| 0.0 | 0.1 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 0.6 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 2.4 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 2.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.2 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.6 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.5 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.6 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.2 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0015786 | UDP-glucose transport(GO:0015786) |
| 0.0 | 0.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 1.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.5 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 6.4 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.1 | GO:0072401 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 3.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.8 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.4 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.5 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:0046325 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.6 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 | 0.1 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.3 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:2000736 | regulation of stem cell differentiation(GO:2000736) |
| 0.0 | 0.7 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.4 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.1 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.1 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.4 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.6 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 2.7 | 8.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 2.3 | 6.8 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 2.0 | 8.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 1.8 | 8.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 1.4 | 4.2 | GO:0035101 | FACT complex(GO:0035101) |
| 1.1 | 5.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 1.1 | 5.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 1.1 | 6.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 1.0 | 8.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 1.0 | 10.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 1.0 | 3.8 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 1.0 | 4.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.9 | 6.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.9 | 14.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.9 | 2.6 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.8 | 2.4 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.8 | 8.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.8 | 5.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.7 | 9.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.7 | 4.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.6 | 4.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.6 | 6.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.5 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 1.4 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.5 | 1.4 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.5 | 1.9 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.4 | 4.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 8.7 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.4 | 3.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 4.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.4 | 2.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.4 | 9.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 2.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.4 | 2.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 2.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.4 | 3.7 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.4 | 1.8 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.4 | 6.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 1.0 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.3 | 1.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 3.0 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 2.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.3 | 14.6 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.3 | 1.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.3 | 7.5 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.3 | 2.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 1.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 2.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 1.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.2 | 4.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.2 | 2.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 9.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 0.8 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 1.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 6.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.2 | 2.6 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.2 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 2.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 0.6 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 3.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 1.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 2.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.2 | 1.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.2 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.2 | 2.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.7 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.8 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 1.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.2 | 3.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.2 | 1.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.2 | 0.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.3 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.9 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.6 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.5 | GO:0033202 | DNA helicase complex(GO:0033202) |
| 0.1 | 0.6 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 1.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 4.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 7.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 3.2 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 6.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 10.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.8 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 8.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 7.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 31.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 4.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 12.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.6 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 2.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 2.8 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.9 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.6 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.0 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.7 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 1.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 7.7 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 12.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.8 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 2.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.1 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 1.1 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 2.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.8 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 24.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 3.6 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 8.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 4.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 6.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 3.1 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 2.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.8 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 9.2 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 3.3 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 1.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 8.1 | GO:0030529 | intracellular ribonucleoprotein complex(GO:0030529) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 8.0 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 3.8 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.6 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 3.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0000800 | lateral element(GO:0000800) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 1.8 | 7.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 1.8 | 8.8 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 1.6 | 6.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.5 | 4.5 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 1.5 | 7.4 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 1.3 | 8.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 1.2 | 6.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 1.2 | 8.7 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 1.2 | 3.6 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 1.2 | 4.8 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.0 | 4.2 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 1.0 | 4.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.9 | 2.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.9 | 5.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.9 | 6.9 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.9 | 6.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.8 | 5.6 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.8 | 3.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.7 | 5.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.7 | 5.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.7 | 2.9 | GO:0035851 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.7 | 2.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.7 | 8.6 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.7 | 8.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.7 | 6.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.7 | 32.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.7 | 4.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.6 | 2.6 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.6 | 16.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.6 | 1.8 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 3.0 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.6 | 3.5 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.6 | 3.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.5 | 2.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.5 | 7.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.5 | 3.7 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.5 | 1.5 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.5 | 5.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.5 | 3.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.5 | 1.4 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.5 | 2.3 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.4 | 5.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 1.3 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 4.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 1.7 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.4 | 10.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.4 | 1.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 5.4 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.4 | 1.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 1.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.4 | 14.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 4.8 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 1.4 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 1.3 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.3 | 7.0 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.3 | 0.9 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 0.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.3 | 1.2 | GO:0038100 | nodal binding(GO:0038100) |
| 0.3 | 2.6 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 2.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 0.8 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.3 | 2.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 1.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 3.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.3 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 0.7 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.2 | 1.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.7 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.2 | 6.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.2 | 15.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.0 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.2 | 1.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.2 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 7.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 9.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 11.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.2 | 1.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 1.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 8.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 1.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 15.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.2 | 1.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 2.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 4.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.2 | 2.7 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 1.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 1.0 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.2 | 7.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 1.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 1.3 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.2 | 5.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.2 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 1.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.2 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 1.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 4.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 4.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 0.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 2.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 0.5 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 10.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 3.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 2.5 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 1.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 2.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 6.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 4.5 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 6.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 2.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 2.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 6.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 2.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 2.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 3.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 3.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 2.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 0.4 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 3.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.7 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.3 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 3.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.0 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.1 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 1.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.7 | GO:1901612 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 2.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 26.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.5 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 4.9 | GO:0070035 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 3.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 4.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 1.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 7.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.1 | 0.7 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.2 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 2.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.6 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.1 | 0.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 14.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.0 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.6 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 2.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.0 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.3 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 2.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 6.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 2.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 9.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 5.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 3.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 11.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 35.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 4.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.3 | 11.5 | PID ATM PATHWAY | ATM pathway |
| 0.2 | 6.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 8.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 4.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 8.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.2 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 7.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 6.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 7.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 7.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.2 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 6.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 3.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 3.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 3.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 5.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 4.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 4.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 2.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 3.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 7.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.6 | 8.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.6 | 19.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.5 | 8.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.5 | 2.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 19.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 9.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.3 | 8.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.3 | 8.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 5.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 33.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.2 | 0.9 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.2 | 21.7 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.2 | 1.4 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.2 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 4.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 6.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.2 | 4.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 21.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 1.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 11.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 3.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.2 | 2.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 7.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 14.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.3 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.1 | 1.7 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 4.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 8.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 4.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 3.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 2.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.8 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 0.7 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 4.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 8.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 3.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.9 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 7.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.9 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.3 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 3.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 10.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME HEPARAN SULFATE HEPARIN HS GAG METABOLISM | Genes involved in Heparan sulfate/heparin (HS-GAG) metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME PHOSPHOLIPID METABOLISM | Genes involved in Phospholipid metabolism |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.2 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.3 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |