Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfap2b
Z-value: 1.12
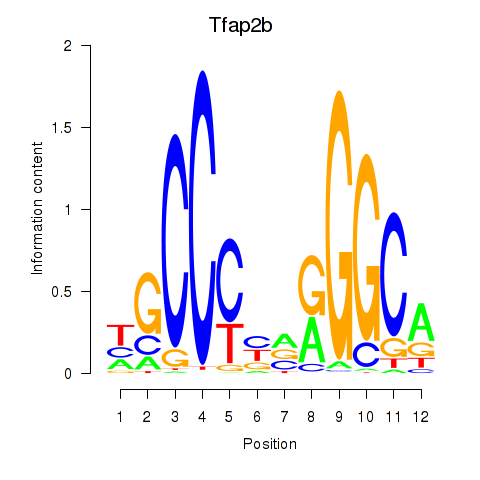
Transcription factors associated with Tfap2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2b
|
ENSMUSG00000025927.7 | transcription factor AP-2 beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2b | mm10_v2_chr1_+_19212054_19212106 | 0.37 | 2.6e-02 | Click! |
Activity profile of Tfap2b motif
Sorted Z-values of Tfap2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102365111 | 6.20 |
ENSMUST00000006749.9
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr12_-_76709997 | 5.61 |
ENSMUST00000166101.1
|
Sptb
|
spectrin beta, erythrocytic |
| chr4_-_140774196 | 5.11 |
ENSMUST00000026381.6
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr3_-_84480419 | 4.55 |
ENSMUST00000107689.1
|
Fhdc1
|
FH2 domain containing 1 |
| chr2_-_131160006 | 4.02 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr7_-_45526146 | 2.46 |
ENSMUST00000167273.1
ENSMUST00000042105.8 |
Ppp1r15a
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A |
| chr12_+_102129019 | 2.34 |
ENSMUST00000079020.4
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr3_-_132950043 | 2.33 |
ENSMUST00000117164.1
ENSMUST00000093971.4 ENSMUST00000042729.9 ENSMUST00000042744.9 ENSMUST00000117811.1 |
Npnt
|
nephronectin |
| chr11_-_70255329 | 2.25 |
ENSMUST00000108574.2
ENSMUST00000000329.2 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr11_+_35121126 | 2.04 |
ENSMUST00000069837.3
|
Slit3
|
slit homolog 3 (Drosophila) |
| chr17_+_33555719 | 2.03 |
ENSMUST00000087605.5
ENSMUST00000174695.1 |
Myo1f
|
myosin IF |
| chr3_-_137981523 | 2.01 |
ENSMUST00000136613.1
ENSMUST00000029806.6 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr7_-_98162318 | 1.96 |
ENSMUST00000107112.1
|
Capn5
|
calpain 5 |
| chr1_-_96872165 | 1.95 |
ENSMUST00000071985.4
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr8_+_79028317 | 1.94 |
ENSMUST00000087927.4
ENSMUST00000098614.2 |
Zfp827
|
zinc finger protein 827 |
| chr2_+_119742306 | 1.89 |
ENSMUST00000028758.7
|
Itpka
|
inositol 1,4,5-trisphosphate 3-kinase A |
| chr6_-_124733067 | 1.86 |
ENSMUST00000173647.1
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr15_-_97831460 | 1.85 |
ENSMUST00000079838.7
ENSMUST00000118294.1 |
Hdac7
|
histone deacetylase 7 |
| chr17_+_34914459 | 1.85 |
ENSMUST00000007249.8
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr11_-_99493112 | 1.81 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr7_-_80387935 | 1.78 |
ENSMUST00000080932.6
|
Fes
|
feline sarcoma oncogene |
| chrX_+_49470450 | 1.76 |
ENSMUST00000114904.3
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr17_-_25952565 | 1.76 |
ENSMUST00000162431.1
|
A930017K11Rik
|
RIKEN cDNA A930017K11 gene |
| chrX_+_49470555 | 1.73 |
ENSMUST00000042444.6
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr12_+_102128718 | 1.71 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr7_-_142969238 | 1.70 |
ENSMUST00000009392.4
ENSMUST00000121862.1 |
Ascl2
|
achaete-scute complex homolog 2 (Drosophila) |
| chrX_-_104671048 | 1.68 |
ENSMUST00000042070.5
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr6_-_37299950 | 1.66 |
ENSMUST00000101532.3
|
Dgki
|
diacylglycerol kinase, iota |
| chr7_+_25686994 | 1.50 |
ENSMUST00000002678.9
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr16_-_22163299 | 1.48 |
ENSMUST00000100052.4
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr19_+_38055002 | 1.45 |
ENSMUST00000096096.4
ENSMUST00000116506.1 ENSMUST00000169673.1 |
Cep55
|
centrosomal protein 55 |
| chrX_+_159627534 | 1.45 |
ENSMUST00000073094.3
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr4_-_136956784 | 1.43 |
ENSMUST00000030420.8
|
Epha8
|
Eph receptor A8 |
| chr5_-_24329556 | 1.42 |
ENSMUST00000115098.2
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr8_+_105297663 | 1.38 |
ENSMUST00000015003.8
|
E2f4
|
E2F transcription factor 4 |
| chr14_-_67715585 | 1.36 |
ENSMUST00000163100.1
ENSMUST00000132705.1 ENSMUST00000124045.1 |
Cdca2
|
cell division cycle associated 2 |
| chr9_-_14751987 | 1.35 |
ENSMUST00000061498.5
|
Fut4
|
fucosyltransferase 4 |
| chr7_+_29134971 | 1.34 |
ENSMUST00000160194.1
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr8_+_84723003 | 1.32 |
ENSMUST00000098571.4
|
G430095P16Rik
|
RIKEN cDNA G430095P16 gene |
| chr2_-_164404606 | 1.29 |
ENSMUST00000109359.1
ENSMUST00000109358.1 ENSMUST00000103103.3 |
Matn4
|
matrilin 4 |
| chr4_-_133967235 | 1.29 |
ENSMUST00000123234.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_+_127169131 | 1.27 |
ENSMUST00000046659.7
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr13_+_48662989 | 1.25 |
ENSMUST00000021813.4
|
Barx1
|
BarH-like homeobox 1 |
| chr5_-_115300912 | 1.23 |
ENSMUST00000112090.1
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr2_-_127729872 | 1.22 |
ENSMUST00000028856.2
|
Mall
|
mal, T cell differentiation protein-like |
| chr5_-_115300957 | 1.15 |
ENSMUST00000009157.3
|
Dynll1
|
dynein light chain LC8-type 1 |
| chr12_+_85686648 | 1.15 |
ENSMUST00000040536.5
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr9_+_119052863 | 1.11 |
ENSMUST00000131647.1
|
Vill
|
villin-like |
| chr3_-_65529355 | 1.09 |
ENSMUST00000099076.3
|
4931440P22Rik
|
RIKEN cDNA 4931440P22 gene |
| chr4_+_46450892 | 1.09 |
ENSMUST00000102926.4
|
Anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr4_-_133967296 | 1.08 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr18_+_60774675 | 1.08 |
ENSMUST00000118551.1
|
Rps14
|
ribosomal protein S14 |
| chr2_+_91945703 | 1.08 |
ENSMUST00000178895.1
|
Gm9821
|
predicted gene 9821 |
| chr1_+_74791516 | 1.07 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr18_+_60774510 | 1.04 |
ENSMUST00000025511.3
|
Rps14
|
ribosomal protein S14 |
| chr2_-_181693810 | 1.04 |
ENSMUST00000108776.1
ENSMUST00000108771.1 ENSMUST00000108779.1 ENSMUST00000108769.1 ENSMUST00000108772.1 ENSMUST00000002532.2 |
Rgs19
|
regulator of G-protein signaling 19 |
| chr2_+_148395369 | 1.01 |
ENSMUST00000109962.2
ENSMUST00000047292.2 |
Sstr4
|
somatostatin receptor 4 |
| chr7_+_44246722 | 0.99 |
ENSMUST00000055858.7
ENSMUST00000107949.1 ENSMUST00000107950.2 ENSMUST00000107948.1 ENSMUST00000084937.4 ENSMUST00000146155.1 |
2410002F23Rik
|
RIKEN cDNA 2410002F23 gene |
| chr9_+_119052770 | 0.99 |
ENSMUST00000051386.6
ENSMUST00000074734.6 |
Vill
|
villin-like |
| chrX_+_159627265 | 0.98 |
ENSMUST00000112456.2
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr2_-_30830333 | 0.98 |
ENSMUST00000041726.3
|
Asb6
|
ankyrin repeat and SOCS box-containing 6 |
| chr16_-_20425881 | 0.97 |
ENSMUST00000077867.3
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr7_+_24636566 | 0.97 |
ENSMUST00000080718.4
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr15_+_99579054 | 0.96 |
ENSMUST00000023752.4
|
Aqp2
|
aquaporin 2 |
| chr17_+_43016536 | 0.95 |
ENSMUST00000024708.4
|
Tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr2_+_25289899 | 0.95 |
ENSMUST00000028337.6
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr11_+_69655314 | 0.94 |
ENSMUST00000047373.5
|
Sox15
|
SRY-box containing gene 15 |
| chr1_-_136234113 | 0.93 |
ENSMUST00000120339.1
ENSMUST00000048668.8 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr9_-_57262591 | 0.93 |
ENSMUST00000034846.5
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr6_+_91515928 | 0.92 |
ENSMUST00000040607.4
|
Lsm3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr7_+_29134854 | 0.91 |
ENSMUST00000161522.1
ENSMUST00000159975.1 ENSMUST00000032811.5 ENSMUST00000094617.4 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr11_+_69965396 | 0.91 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr17_-_35700520 | 0.91 |
ENSMUST00000119825.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr4_-_133967893 | 0.90 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_58658181 | 0.90 |
ENSMUST00000168747.1
|
Atp10a
|
ATPase, class V, type 10A |
| chr4_+_109978004 | 0.89 |
ENSMUST00000061187.3
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2 |
| chr1_+_34801704 | 0.89 |
ENSMUST00000047664.9
|
Arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr18_+_60925612 | 0.87 |
ENSMUST00000102888.3
ENSMUST00000025519.4 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr7_+_141447645 | 0.87 |
ENSMUST00000106004.1
ENSMUST00000106003.1 |
Rplp2
|
ribosomal protein, large P2 |
| chr11_+_32533290 | 0.83 |
ENSMUST00000102821.3
|
Stk10
|
serine/threonine kinase 10 |
| chr11_-_96977660 | 0.82 |
ENSMUST00000107626.1
ENSMUST00000107624.1 |
Sp2
|
Sp2 transcription factor |
| chr7_-_126447642 | 0.81 |
ENSMUST00000146973.1
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr9_+_37489281 | 0.80 |
ENSMUST00000048604.6
|
Msantd2
|
Myb/SANT-like DNA-binding domain containing 2 |
| chr17_+_34590162 | 0.79 |
ENSMUST00000173772.1
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr11_-_94242701 | 0.78 |
ENSMUST00000061469.3
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr17_+_34589799 | 0.78 |
ENSMUST00000038244.8
|
Gpsm3
|
G-protein signalling modulator 3 (AGS3-like, C. elegans) |
| chr1_-_135105210 | 0.77 |
ENSMUST00000044828.7
|
Lgr6
|
leucine-rich repeat-containing G protein-coupled receptor 6 |
| chr8_+_105605220 | 0.76 |
ENSMUST00000043531.8
|
Fam65a
|
family with sequence similarity 65, member A |
| chr5_+_52582320 | 0.75 |
ENSMUST00000177881.1
|
Gm5866
|
predicted gene 5866 |
| chr12_-_111672290 | 0.73 |
ENSMUST00000001304.7
|
Ckb
|
creatine kinase, brain |
| chr10_-_59951753 | 0.73 |
ENSMUST00000020308.3
|
Ddit4
|
DNA-damage-inducible transcript 4 |
| chr2_+_25262589 | 0.73 |
ENSMUST00000114336.3
|
Tprn
|
taperin |
| chr11_+_121259983 | 0.71 |
ENSMUST00000106113.1
|
Foxk2
|
forkhead box K2 |
| chr11_+_5861886 | 0.69 |
ENSMUST00000102923.3
|
Aebp1
|
AE binding protein 1 |
| chr1_+_55406163 | 0.69 |
ENSMUST00000042986.8
|
Plcl1
|
phospholipase C-like 1 |
| chr2_+_158610731 | 0.69 |
ENSMUST00000045738.4
|
Slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr15_-_79834224 | 0.68 |
ENSMUST00000109623.1
ENSMUST00000109625.1 ENSMUST00000023060.6 ENSMUST00000089299.5 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chr11_+_98386450 | 0.67 |
ENSMUST00000041301.7
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr5_-_103629279 | 0.66 |
ENSMUST00000031263.1
|
Slc10a6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6 |
| chr1_+_74771886 | 0.66 |
ENSMUST00000006716.6
|
Wnt6
|
wingless-related MMTV integration site 6 |
| chrX_+_9272756 | 0.66 |
ENSMUST00000015486.6
|
Xk
|
Kell blood group precursor (McLeod phenotype) homolog |
| chrX_+_162760427 | 0.65 |
ENSMUST00000112326.1
|
Rbbp7
|
retinoblastoma binding protein 7 |
| chr12_+_108410625 | 0.65 |
ENSMUST00000109857.1
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr19_-_46039621 | 0.65 |
ENSMUST00000056931.7
|
Ldb1
|
LIM domain binding 1 |
| chr13_+_55464237 | 0.65 |
ENSMUST00000046533.7
|
Prr7
|
proline rich 7 (synaptic) |
| chr15_-_99370427 | 0.64 |
ENSMUST00000081224.7
ENSMUST00000120633.1 ENSMUST00000088233.6 |
Fmnl3
|
formin-like 3 |
| chr4_-_133967953 | 0.63 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr12_+_17690793 | 0.63 |
ENSMUST00000071858.3
|
Hpcal1
|
hippocalcin-like 1 |
| chr15_+_100761741 | 0.63 |
ENSMUST00000023776.6
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr1_+_135766085 | 0.62 |
ENSMUST00000038945.5
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr7_-_45466894 | 0.61 |
ENSMUST00000033093.8
|
Bax
|
BCL2-associated X protein |
| chr15_-_79834261 | 0.60 |
ENSMUST00000148358.1
|
Cbx6
|
chromobox 6 |
| chr9_+_54699514 | 0.60 |
ENSMUST00000154690.1
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr15_-_75747922 | 0.60 |
ENSMUST00000062002.4
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr12_-_15816762 | 0.59 |
ENSMUST00000020922.7
|
Trib2
|
tribbles homolog 2 (Drosophila) |
| chr2_-_11502090 | 0.58 |
ENSMUST00000179584.1
ENSMUST00000170196.2 ENSMUST00000171188.2 ENSMUST00000114845.3 ENSMUST00000114844.1 ENSMUST00000100411.2 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr14_+_20674311 | 0.58 |
ENSMUST00000048657.8
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr9_+_54699548 | 0.58 |
ENSMUST00000070070.7
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chrX_+_151047170 | 0.56 |
ENSMUST00000026296.7
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr19_+_4099998 | 0.55 |
ENSMUST00000049658.7
|
Pitpnm1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr7_-_30914327 | 0.55 |
ENSMUST00000040548.7
|
Mag
|
myelin-associated glycoprotein |
| chr11_-_100414829 | 0.55 |
ENSMUST00000066489.6
|
Leprel4
|
leprecan-like 4 |
| chr2_+_155956537 | 0.53 |
ENSMUST00000109619.2
ENSMUST00000039994.7 ENSMUST00000094421.4 ENSMUST00000151569.1 ENSMUST00000109618.1 |
Cep250
|
centrosomal protein 250 |
| chr4_+_109406623 | 0.53 |
ENSMUST00000124209.1
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr2_+_74697663 | 0.52 |
ENSMUST00000059272.8
|
Hoxd9
|
homeobox D9 |
| chr12_+_108410542 | 0.51 |
ENSMUST00000054955.7
|
Eml1
|
echinoderm microtubule associated protein like 1 |
| chr11_-_58168467 | 0.51 |
ENSMUST00000172035.1
ENSMUST00000035604.6 ENSMUST00000102711.2 |
Gemin5
|
gem (nuclear organelle) associated protein 5 |
| chr7_+_45526330 | 0.51 |
ENSMUST00000120985.1
ENSMUST00000051810.8 |
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr14_-_30607808 | 0.50 |
ENSMUST00000112207.1
ENSMUST00000112206.1 ENSMUST00000112202.1 ENSMUST00000112203.1 |
Prkcd
|
protein kinase C, delta |
| chr3_-_57847478 | 0.50 |
ENSMUST00000120289.1
ENSMUST00000066882.8 |
Pfn2
|
profilin 2 |
| chr14_+_79451791 | 0.49 |
ENSMUST00000100359.1
|
Zbtbd6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr8_-_122678072 | 0.49 |
ENSMUST00000006525.7
ENSMUST00000064674.6 |
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr2_-_24475097 | 0.48 |
ENSMUST00000149294.1
ENSMUST00000153535.1 |
Pax8
|
paired box gene 8 |
| chr6_-_124733121 | 0.48 |
ENSMUST00000112484.3
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr2_-_11502067 | 0.48 |
ENSMUST00000028114.6
ENSMUST00000049849.6 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr4_-_133753611 | 0.48 |
ENSMUST00000145664.2
ENSMUST00000105897.3 |
Arid1a
|
AT rich interactive domain 1A (SWI-like) |
| chr10_-_119240006 | 0.48 |
ENSMUST00000020315.6
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr3_+_32817520 | 0.48 |
ENSMUST00000072312.5
ENSMUST00000108228.1 |
Usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr19_+_43440404 | 0.47 |
ENSMUST00000165311.1
|
Cnnm1
|
cyclin M1 |
| chr3_-_27710413 | 0.47 |
ENSMUST00000046157.4
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr4_-_134238372 | 0.44 |
ENSMUST00000030645.8
|
Cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chrX_+_162760388 | 0.43 |
ENSMUST00000033720.5
ENSMUST00000112327.1 |
Rbbp7
|
retinoblastoma binding protein 7 |
| chr9_-_50344981 | 0.42 |
ENSMUST00000076364.4
|
Rpl10-ps3
|
ribosomal protein L10, pseudogene 3 |
| chr17_-_46282991 | 0.42 |
ENSMUST00000180283.1
ENSMUST00000012440.6 ENSMUST00000164342.2 |
Tjap1
|
tight junction associated protein 1 |
| chr2_+_22895583 | 0.41 |
ENSMUST00000152170.1
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr15_+_102966794 | 0.40 |
ENSMUST00000001699.7
|
Hoxc10
|
homeobox C10 |
| chr19_+_44757394 | 0.40 |
ENSMUST00000004340.4
|
Pax2
|
paired box gene 2 |
| chr2_+_27165233 | 0.39 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr7_-_93081027 | 0.38 |
ENSMUST00000098303.1
|
Gm9934
|
predicted gene 9934 |
| chr13_+_44731265 | 0.38 |
ENSMUST00000173246.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_+_24507122 | 0.38 |
ENSMUST00000177205.1
|
Zfp428
|
zinc finger protein 428 |
| chr7_+_141079125 | 0.38 |
ENSMUST00000159375.1
|
Pkp3
|
plakophilin 3 |
| chr12_+_70825492 | 0.38 |
ENSMUST00000057859.7
|
Frmd6
|
FERM domain containing 6 |
| chr2_-_11502025 | 0.37 |
ENSMUST00000114846.2
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr2_+_130406478 | 0.36 |
ENSMUST00000055421.4
|
Tmem239
|
transmembrane 239 |
| chr7_-_126503408 | 0.36 |
ENSMUST00000040202.8
|
Atxn2l
|
ataxin 2-like |
| chr2_-_25580099 | 0.35 |
ENSMUST00000114217.1
|
Gm996
|
predicted gene 996 |
| chr17_+_35841491 | 0.35 |
ENSMUST00000082337.6
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr1_-_14755966 | 0.34 |
ENSMUST00000027062.5
|
Msc
|
musculin |
| chr10_+_39420009 | 0.34 |
ENSMUST00000157009.1
|
Fyn
|
Fyn proto-oncogene |
| chr19_-_6015769 | 0.34 |
ENSMUST00000164843.1
|
Capn1
|
calpain 1 |
| chr4_+_123183722 | 0.34 |
ENSMUST00000152194.1
|
Hpcal4
|
hippocalcin-like 4 |
| chr2_-_131086765 | 0.33 |
ENSMUST00000028794.3
ENSMUST00000110227.1 ENSMUST00000110226.1 |
Siglec1
|
sialic acid binding Ig-like lectin 1, sialoadhesin |
| chr1_+_131750485 | 0.31 |
ENSMUST00000147800.1
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr2_-_92024502 | 0.30 |
ENSMUST00000028663.4
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr15_-_101924725 | 0.29 |
ENSMUST00000023797.6
|
Krt4
|
keratin 4 |
| chr15_-_79834323 | 0.28 |
ENSMUST00000177316.2
ENSMUST00000175858.2 |
Nptxr
|
neuronal pentraxin receptor |
| chr10_-_127211528 | 0.28 |
ENSMUST00000013970.7
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr13_-_9878998 | 0.28 |
ENSMUST00000063093.9
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr19_+_6364557 | 0.28 |
ENSMUST00000155973.1
|
Sf1
|
splicing factor 1 |
| chr16_-_92358874 | 0.28 |
ENSMUST00000166707.1
|
Kcne1
|
potassium voltage-gated channel, Isk-related subfamily, member 1 |
| chr19_-_53038534 | 0.28 |
ENSMUST00000183274.1
ENSMUST00000182097.1 ENSMUST00000069988.8 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr7_+_25282179 | 0.27 |
ENSMUST00000163320.1
ENSMUST00000005578.6 |
Cic
|
capicua homolog (Drosophila) |
| chr2_-_179976646 | 0.26 |
ENSMUST00000041618.7
|
Taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_-_172043466 | 0.25 |
ENSMUST00000087950.3
|
Cbln4
|
cerebellin 4 precursor protein |
| chr6_+_88465409 | 0.25 |
ENSMUST00000032165.9
|
Ruvbl1
|
RuvB-like protein 1 |
| chr4_+_133176336 | 0.25 |
ENSMUST00000105912.1
|
Wasf2
|
WAS protein family, member 2 |
| chr17_-_32284715 | 0.25 |
ENSMUST00000127893.1
|
Brd4
|
bromodomain containing 4 |
| chr15_+_103018936 | 0.24 |
ENSMUST00000165375.1
|
Hoxc4
|
homeobox C4 |
| chr5_+_30913398 | 0.24 |
ENSMUST00000031055.5
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr4_+_8690399 | 0.23 |
ENSMUST00000127476.1
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr15_-_38300693 | 0.23 |
ENSMUST00000074043.5
|
Klf10
|
Kruppel-like factor 10 |
| chr13_+_12702362 | 0.23 |
ENSMUST00000104944.2
|
Gm2399
|
predicted gene 2399 |
| chr17_+_34898931 | 0.22 |
ENSMUST00000097342.3
ENSMUST00000013931.5 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr17_-_12992188 | 0.22 |
ENSMUST00000159986.1
|
Wtap
|
Wilms' tumour 1-associating protein |
| chr6_-_24956106 | 0.21 |
ENSMUST00000127247.2
|
Tmem229a
|
transmembrane protein 229A |
| chr8_+_105413614 | 0.21 |
ENSMUST00000109355.2
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr17_-_70849644 | 0.21 |
ENSMUST00000134654.1
ENSMUST00000172229.1 ENSMUST00000127719.1 |
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr16_+_20629842 | 0.21 |
ENSMUST00000122306.1
ENSMUST00000133344.1 |
Ece2
|
endothelin converting enzyme 2 |
| chr1_-_143702832 | 0.21 |
ENSMUST00000018337.7
|
Cdc73
|
cell division cycle 73, Paf1/RNA polymerase II complex component |
| chr15_-_11905609 | 0.20 |
ENSMUST00000066529.3
|
Npr3
|
natriuretic peptide receptor 3 |
| chr11_-_95146263 | 0.20 |
ENSMUST00000021241.6
|
Dlx4
|
distal-less homeobox 4 |
| chr11_+_102189620 | 0.20 |
ENSMUST00000070334.3
ENSMUST00000078975.7 |
G6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chrX_+_73757069 | 0.20 |
ENSMUST00000002079.6
|
Plxnb3
|
plexin B3 |
| chr4_+_43875524 | 0.20 |
ENSMUST00000030198.6
|
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr19_+_6975048 | 0.17 |
ENSMUST00000070850.6
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr13_+_44731281 | 0.16 |
ENSMUST00000174086.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr3_+_89831352 | 0.16 |
ENSMUST00000050401.5
|
She
|
src homology 2 domain-containing transforming protein E |
| chr8_+_104540800 | 0.16 |
ENSMUST00000056051.4
|
Car7
|
carbonic anhydrase 7 |
| chr10_-_7663245 | 0.16 |
ENSMUST00000163085.1
ENSMUST00000159917.1 |
Pcmt1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.8 | 2.5 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.6 | 1.8 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.6 | 2.3 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.5 | 1.5 | GO:0044413 | CD4-positive, CD25-positive, alpha-beta regulatory T cell lineage commitment(GO:0002362) evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.5 | 2.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.5 | 2.3 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.4 | 2.0 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.4 | 1.4 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.0 | GO:0071462 | cellular response to mercury ion(GO:0071288) cellular response to water stimulus(GO:0071462) |
| 0.3 | 0.9 | GO:0048627 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) myoblast development(GO:0048627) |
| 0.3 | 0.6 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.3 | 1.7 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 1.1 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.3 | 0.8 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) maintenance of mitochondrion location(GO:0051659) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 0.8 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.2 | 1.0 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 | 6.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 0.7 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.2 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.2 | 0.6 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.2 | 4.0 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.2 | 1.7 | GO:0046959 | habituation(GO:0046959) |
| 0.2 | 0.6 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.2 | 1.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.5 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.2 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 2.0 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.2 | 1.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.9 | GO:2000594 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.1 | 0.9 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 1.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 2.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 7.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.7 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 0.5 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 3.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 0.4 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 1.9 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.3 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.7 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 1.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 1.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.1 | 0.5 | GO:0060697 | glucosylceramide catabolic process(GO:0006680) positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 1.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 1.7 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.1 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.7 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.9 | GO:0070423 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 1.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.7 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.3 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 1.5 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0036166 | phenotypic switching(GO:0036166) cellular response to cocaine(GO:0071314) |
| 0.1 | 1.6 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.1 | 0.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) atrioventricular canal development(GO:0036302) |
| 0.1 | 0.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.1 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.1 | 0.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.1 | 0.2 | GO:1902460 | transforming growth factor beta activation(GO:0036363) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.9 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 1.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.4 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 1.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.3 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.5 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.3 | GO:0010756 | box C/D snoRNP assembly(GO:0000492) positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 1.3 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 1.8 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 | 0.6 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 1.2 | GO:0030815 | negative regulation of cAMP metabolic process(GO:0030815) |
| 0.0 | 0.2 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 1.1 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.8 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 2.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.8 | GO:0048678 | response to axon injury(GO:0048678) |
| 0.0 | 0.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.7 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.3 | 2.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.3 | 2.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.2 | 0.9 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.2 | 1.0 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 0.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0034774 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 1.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 1.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 1.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.4 | GO:0036396 | MIS complex(GO:0036396) |
| 0.1 | 0.4 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 0.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.3 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.1 | 6.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 1.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 2.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 2.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 1.9 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.7 | 4.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.6 | 2.3 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.4 | 1.5 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 6.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 1.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.3 | 0.9 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 0.3 | 2.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.3 | 1.8 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 2.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.2 | 1.9 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 1.0 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 2.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 1.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 2.4 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.7 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 1.0 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 5.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.5 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.4 | GO:0000010 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 2.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 1.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 2.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.8 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.2 | GO:0034437 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.1 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.7 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.9 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.9 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 3.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.2 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.3 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 3.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 3.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 2.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 5.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 3.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 11.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.0 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.1 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 2.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 1.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |