Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfap2a
Z-value: 1.05
Transcription factors associated with Tfap2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2a
|
ENSMUSG00000021359.9 | transcription factor AP-2, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfap2a | mm10_v2_chr13_-_40733768_40733836 | 0.28 | 9.6e-02 | Click! |
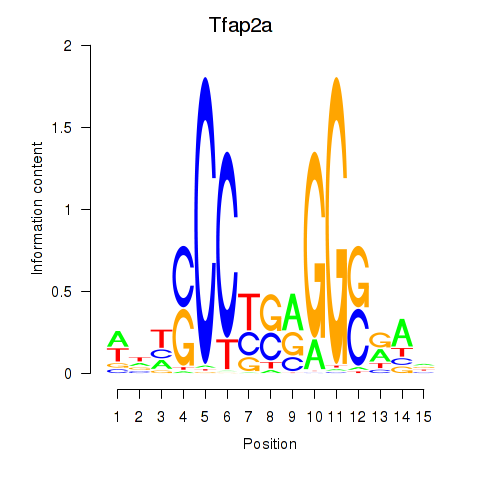
Activity profile of Tfap2a motif
Sorted Z-values of Tfap2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_82946898 | 3.46 |
ENSMUST00000113980.3
|
M1ap
|
meiosis 1 associated protein |
| chr3_-_84480419 | 2.93 |
ENSMUST00000107689.1
|
Fhdc1
|
FH2 domain containing 1 |
| chr10_+_75571522 | 2.53 |
ENSMUST00000143226.1
ENSMUST00000124259.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr5_-_43981757 | 2.39 |
ENSMUST00000061299.7
|
Fgfbp1
|
fibroblast growth factor binding protein 1 |
| chr9_-_103480328 | 2.27 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr9_-_44454757 | 2.19 |
ENSMUST00000047740.2
|
Upk2
|
uroplakin 2 |
| chr11_-_100135928 | 2.17 |
ENSMUST00000107411.2
|
Krt15
|
keratin 15 |
| chr10_+_75566257 | 1.95 |
ENSMUST00000129232.1
ENSMUST00000143792.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr12_-_8539545 | 1.81 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr7_+_24636566 | 1.76 |
ENSMUST00000080718.4
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr11_+_32296489 | 1.72 |
ENSMUST00000093207.3
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr15_-_76669811 | 1.71 |
ENSMUST00000037824.4
|
Foxh1
|
forkhead box H1 |
| chr17_+_48359891 | 1.69 |
ENSMUST00000024792.6
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr10_+_100015817 | 1.69 |
ENSMUST00000130190.1
ENSMUST00000020129.7 |
Kitl
|
kit ligand |
| chr9_+_110857958 | 1.65 |
ENSMUST00000051097.1
|
Prss50
|
protease, serine, 50 |
| chr12_+_109452833 | 1.61 |
ENSMUST00000056110.8
|
Dlk1
|
delta-like 1 homolog (Drosophila) |
| chr4_-_136956784 | 1.58 |
ENSMUST00000030420.8
|
Epha8
|
Eph receptor A8 |
| chrX_+_93675088 | 1.57 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr11_+_80089385 | 1.53 |
ENSMUST00000108239.1
ENSMUST00000017694.5 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr7_-_131322292 | 1.51 |
ENSMUST00000046611.7
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr1_+_172555932 | 1.45 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr5_-_24030297 | 1.41 |
ENSMUST00000101513.2
|
Fam126a
|
family with sequence similarity 126, member A |
| chr8_-_111691002 | 1.40 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr11_-_102897123 | 1.40 |
ENSMUST00000067444.3
|
Gfap
|
glial fibrillary acidic protein |
| chr11_-_102897146 | 1.40 |
ENSMUST00000077902.4
|
Gfap
|
glial fibrillary acidic protein |
| chr4_-_140774196 | 1.39 |
ENSMUST00000026381.6
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr2_+_154190806 | 1.38 |
ENSMUST00000081816.4
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr11_+_95010277 | 1.36 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr15_-_103252810 | 1.34 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_+_79255500 | 1.33 |
ENSMUST00000099972.4
|
Itga4
|
integrin alpha 4 |
| chr17_-_87797994 | 1.33 |
ENSMUST00000055221.7
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr7_-_24545994 | 1.31 |
ENSMUST00000011776.6
|
Pinlyp
|
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr1_-_96872165 | 1.30 |
ENSMUST00000071985.4
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr19_+_7268296 | 1.30 |
ENSMUST00000066646.4
|
Rcor2
|
REST corepressor 2 |
| chr7_+_30787897 | 1.29 |
ENSMUST00000098559.1
|
Krtdap
|
keratinocyte differentiation associated protein |
| chr13_-_53286052 | 1.20 |
ENSMUST00000021918.8
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr9_-_123968683 | 1.19 |
ENSMUST00000026911.4
|
Ccr1
|
chemokine (C-C motif) receptor 1 |
| chr2_-_131160006 | 1.19 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr15_+_79892436 | 1.12 |
ENSMUST00000175752.1
ENSMUST00000176325.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr2_+_103424120 | 1.11 |
ENSMUST00000171693.1
|
Elf5
|
E74-like factor 5 |
| chr12_+_109544498 | 1.09 |
ENSMUST00000126289.1
|
Meg3
|
maternally expressed 3 |
| chr4_-_127330799 | 1.08 |
ENSMUST00000046532.3
|
Gjb3
|
gap junction protein, beta 3 |
| chr15_-_89425856 | 1.08 |
ENSMUST00000109313.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr16_-_22657182 | 1.08 |
ENSMUST00000023578.7
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr15_+_79892397 | 1.06 |
ENSMUST00000175714.1
ENSMUST00000109620.3 ENSMUST00000165537.1 |
Apobec3
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 3 |
| chr2_+_103424056 | 1.03 |
ENSMUST00000028609.7
|
Elf5
|
E74-like factor 5 |
| chrX_-_136215443 | 1.02 |
ENSMUST00000113120.1
ENSMUST00000113118.1 ENSMUST00000058125.8 |
Bex1
|
brain expressed gene 1 |
| chr10_+_79879614 | 1.00 |
ENSMUST00000006679.8
|
Prtn3
|
proteinase 3 |
| chr10_+_79881023 | 0.99 |
ENSMUST00000166201.1
|
Prtn3
|
proteinase 3 |
| chr9_+_92542223 | 0.98 |
ENSMUST00000070522.7
ENSMUST00000160359.1 |
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr13_+_73467197 | 0.98 |
ENSMUST00000022099.8
|
Lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr11_+_116532441 | 0.98 |
ENSMUST00000106386.1
ENSMUST00000145737.1 ENSMUST00000155102.1 ENSMUST00000063446.6 |
Sphk1
|
sphingosine kinase 1 |
| chr14_+_54640952 | 0.94 |
ENSMUST00000169818.2
|
Gm17606
|
predicted gene, 17606 |
| chr13_-_98206151 | 0.92 |
ENSMUST00000109426.1
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr16_+_32608973 | 0.92 |
ENSMUST00000120680.1
|
Tfrc
|
transferrin receptor |
| chr11_+_67078293 | 0.91 |
ENSMUST00000108689.1
ENSMUST00000165221.1 ENSMUST00000007301.7 |
Myh3
|
myosin, heavy polypeptide 3, skeletal muscle, embryonic |
| chr8_+_120002720 | 0.90 |
ENSMUST00000108972.3
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr16_-_22657165 | 0.90 |
ENSMUST00000089925.3
|
Dgkg
|
diacylglycerol kinase, gamma |
| chrX_+_48519245 | 0.90 |
ENSMUST00000033430.2
|
Rab33a
|
RAB33A, member of RAS oncogene family |
| chr19_-_45816007 | 0.88 |
ENSMUST00000079431.3
ENSMUST00000026247.6 ENSMUST00000162528.2 |
Kcnip2
|
Kv channel-interacting protein 2 |
| chr2_+_129228022 | 0.88 |
ENSMUST00000148548.1
|
A730036I17Rik
|
RIKEN cDNA A730036I17 gene |
| chr8_+_70493156 | 0.85 |
ENSMUST00000008032.7
|
Crlf1
|
cytokine receptor-like factor 1 |
| chrX_+_49470450 | 0.84 |
ENSMUST00000114904.3
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr17_-_36867187 | 0.84 |
ENSMUST00000025329.6
ENSMUST00000174195.1 |
Trim15
|
tripartite motif-containing 15 |
| chr3_-_137981523 | 0.82 |
ENSMUST00000136613.1
ENSMUST00000029806.6 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr17_-_45474839 | 0.81 |
ENSMUST00000024731.8
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr1_+_136131382 | 0.81 |
ENSMUST00000075164.4
|
Kif21b
|
kinesin family member 21B |
| chr5_+_33658550 | 0.81 |
ENSMUST00000152847.1
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_-_163918683 | 0.79 |
ENSMUST00000044734.2
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr5_-_24030649 | 0.79 |
ENSMUST00000030849.6
|
Fam126a
|
family with sequence similarity 126, member A |
| chr2_-_24935054 | 0.78 |
ENSMUST00000132074.1
|
Arrdc1
|
arrestin domain containing 1 |
| chr12_-_115964196 | 0.78 |
ENSMUST00000103550.2
|
Ighv1-83
|
immunoglobulin heavy variable 1-83 |
| chr7_-_4752972 | 0.77 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chrX_+_49470555 | 0.77 |
ENSMUST00000042444.6
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr8_+_84701430 | 0.77 |
ENSMUST00000037165.4
|
Lyl1
|
lymphoblastomic leukemia 1 |
| chr2_+_91945703 | 0.77 |
ENSMUST00000178895.1
|
Gm9821
|
predicted gene 9821 |
| chr9_+_54698859 | 0.76 |
ENSMUST00000120452.1
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr13_-_49147931 | 0.76 |
ENSMUST00000162581.1
ENSMUST00000110097.2 ENSMUST00000049265.8 ENSMUST00000035538.6 ENSMUST00000110096.1 ENSMUST00000091623.3 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr17_-_24443093 | 0.75 |
ENSMUST00000088506.5
|
Dnase1l2
|
deoxyribonuclease 1-like 2 |
| chr2_-_160912292 | 0.74 |
ENSMUST00000109454.1
ENSMUST00000057169.4 |
Emilin3
|
elastin microfibril interfacer 3 |
| chr5_-_137072254 | 0.72 |
ENSMUST00000077523.3
ENSMUST00000041388.4 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr3_+_152165374 | 0.70 |
ENSMUST00000181854.1
|
D630002J18Rik
|
RIKEN cDNA D630002J18 gene |
| chr9_-_97111117 | 0.69 |
ENSMUST00000085206.4
|
Slc25a36
|
solute carrier family 25, member 36 |
| chr11_+_32283511 | 0.69 |
ENSMUST00000093209.3
|
Hba-a1
|
hemoglobin alpha, adult chain 1 |
| chr7_-_3677509 | 0.69 |
ENSMUST00000038743.8
|
Tmc4
|
transmembrane channel-like gene family 4 |
| chr1_-_75278345 | 0.68 |
ENSMUST00000039534.4
|
Resp18
|
regulated endocrine-specific protein 18 |
| chr12_-_109068173 | 0.68 |
ENSMUST00000073156.7
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr8_-_105637403 | 0.68 |
ENSMUST00000182046.1
|
Gm5914
|
predicted gene 5914 |
| chr6_-_60828889 | 0.67 |
ENSMUST00000114268.3
|
Snca
|
synuclein, alpha |
| chr10_+_11281583 | 0.67 |
ENSMUST00000070300.4
|
Fbxo30
|
F-box protein 30 |
| chr19_+_53600377 | 0.67 |
ENSMUST00000025930.9
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr6_+_90619241 | 0.67 |
ENSMUST00000032177.8
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr19_+_34922351 | 0.67 |
ENSMUST00000087341.5
|
Kif20b
|
kinesin family member 20B |
| chr11_-_61719946 | 0.66 |
ENSMUST00000151780.1
ENSMUST00000148584.1 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr1_-_160153571 | 0.66 |
ENSMUST00000039178.5
|
Tnn
|
tenascin N |
| chr2_+_29890534 | 0.65 |
ENSMUST00000113764.3
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr3_-_88384433 | 0.65 |
ENSMUST00000076048.4
|
Bglap
|
bone gamma carboxyglutamate protein |
| chr11_+_69913888 | 0.64 |
ENSMUST00000072581.2
ENSMUST00000116358.1 |
Gps2
|
G protein pathway suppressor 2 |
| chr12_-_76822510 | 0.64 |
ENSMUST00000021459.7
|
Rab15
|
RAB15, member RAS oncogene family |
| chr6_-_124733067 | 0.64 |
ENSMUST00000173647.1
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr5_+_33658567 | 0.63 |
ENSMUST00000114426.3
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr6_-_82774448 | 0.63 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr17_+_43568641 | 0.63 |
ENSMUST00000169694.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_+_43568475 | 0.62 |
ENSMUST00000167418.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr16_-_4003750 | 0.62 |
ENSMUST00000171658.1
ENSMUST00000171762.1 |
Slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr4_-_154025926 | 0.62 |
ENSMUST00000132541.1
ENSMUST00000143047.1 |
Smim1
|
small integral membrane protein 1 |
| chr8_+_41239718 | 0.62 |
ENSMUST00000045218.7
|
Pcm1
|
pericentriolar material 1 |
| chr10_-_85102487 | 0.62 |
ENSMUST00000059383.6
|
Fhl4
|
four and a half LIM domains 4 |
| chr6_+_141249161 | 0.61 |
ENSMUST00000043259.7
|
Pde3a
|
phosphodiesterase 3A, cGMP inhibited |
| chr2_-_33887862 | 0.61 |
ENSMUST00000041555.3
|
Mvb12b
|
multivesicular body subunit 12B |
| chr3_-_132950043 | 0.61 |
ENSMUST00000117164.1
ENSMUST00000093971.4 ENSMUST00000042729.9 ENSMUST00000042744.9 ENSMUST00000117811.1 |
Npnt
|
nephronectin |
| chr2_-_34754364 | 0.61 |
ENSMUST00000142436.1
ENSMUST00000113099.3 ENSMUST00000028224.8 |
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr7_+_28863831 | 0.61 |
ENSMUST00000138272.1
|
Lgals7
|
lectin, galactose binding, soluble 7 |
| chr19_-_6015152 | 0.60 |
ENSMUST00000025891.8
|
Capn1
|
calpain 1 |
| chr17_-_29237759 | 0.60 |
ENSMUST00000137727.1
ENSMUST00000024805.7 |
Cpne5
|
copine V |
| chr4_+_43383449 | 0.59 |
ENSMUST00000135216.1
ENSMUST00000152322.1 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr3_-_88378699 | 0.59 |
ENSMUST00000098956.2
|
Bglap2
|
bone gamma-carboxyglutamate protein 2 |
| chrX_-_150657392 | 0.58 |
ENSMUST00000151403.2
ENSMUST00000087253.4 ENSMUST00000112709.1 ENSMUST00000163969.1 ENSMUST00000087258.3 |
Tro
|
trophinin |
| chr2_-_121271315 | 0.58 |
ENSMUST00000131245.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr18_+_60925612 | 0.57 |
ENSMUST00000102888.3
ENSMUST00000025519.4 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr10_+_11281304 | 0.57 |
ENSMUST00000129456.1
|
Fbxo30
|
F-box protein 30 |
| chr11_+_98386450 | 0.56 |
ENSMUST00000041301.7
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr11_-_71033462 | 0.56 |
ENSMUST00000156068.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr6_-_50456085 | 0.56 |
ENSMUST00000146341.1
ENSMUST00000071728.4 |
Osbpl3
|
oxysterol binding protein-like 3 |
| chr11_+_63131512 | 0.56 |
ENSMUST00000018361.3
|
Pmp22
|
peripheral myelin protein 22 |
| chr13_+_108214389 | 0.56 |
ENSMUST00000022207.8
|
Elovl7
|
ELOVL family member 7, elongation of long chain fatty acids (yeast) |
| chr17_+_43568269 | 0.56 |
ENSMUST00000024706.5
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr9_+_54699514 | 0.56 |
ENSMUST00000154690.1
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr4_+_149485215 | 0.56 |
ENSMUST00000124413.1
ENSMUST00000141293.1 |
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr16_-_20425881 | 0.55 |
ENSMUST00000077867.3
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr9_+_70679016 | 0.55 |
ENSMUST00000144537.1
|
Adam10
|
a disintegrin and metallopeptidase domain 10 |
| chr16_-_3718105 | 0.55 |
ENSMUST00000023180.7
ENSMUST00000100222.2 |
Mefv
|
Mediterranean fever |
| chr17_-_24443077 | 0.54 |
ENSMUST00000119932.1
|
Dnase1l2
|
deoxyribonuclease 1-like 2 |
| chr16_-_20426375 | 0.54 |
ENSMUST00000079158.6
|
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr2_+_52072823 | 0.54 |
ENSMUST00000112693.2
ENSMUST00000069794.5 |
Rif1
|
Rap1 interacting factor 1 homolog (yeast) |
| chr17_+_27556641 | 0.53 |
ENSMUST00000119486.1
ENSMUST00000118599.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr5_-_67847400 | 0.53 |
ENSMUST00000113652.1
ENSMUST00000113651.1 ENSMUST00000037380.8 |
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr2_+_152911311 | 0.52 |
ENSMUST00000028970.7
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr1_-_193370260 | 0.52 |
ENSMUST00000016323.4
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr2_+_69861638 | 0.52 |
ENSMUST00000112260.1
|
Ssb
|
Sjogren syndrome antigen B |
| chr2_+_5951440 | 0.51 |
ENSMUST00000060092.6
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_+_100227638 | 0.51 |
ENSMUST00000054436.8
|
Pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr12_+_105705970 | 0.51 |
ENSMUST00000040876.5
|
Ak7
|
adenylate kinase 7 |
| chr10_-_81500132 | 0.51 |
ENSMUST00000053646.5
|
S1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr11_-_61720795 | 0.51 |
ENSMUST00000051552.4
|
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr16_+_32608920 | 0.50 |
ENSMUST00000023486.8
|
Tfrc
|
transferrin receptor |
| chr1_-_125913101 | 0.50 |
ENSMUST00000161361.1
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr7_+_29134971 | 0.49 |
ENSMUST00000160194.1
|
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr3_+_33799791 | 0.49 |
ENSMUST00000099153.3
|
Ttc14
|
tetratricopeptide repeat domain 14 |
| chr2_+_103424820 | 0.49 |
ENSMUST00000126290.1
|
Elf5
|
E74-like factor 5 |
| chr19_-_9899450 | 0.49 |
ENSMUST00000025562.7
|
Incenp
|
inner centromere protein |
| chrX_-_107403295 | 0.49 |
ENSMUST00000033591.5
|
Itm2a
|
integral membrane protein 2A |
| chr6_+_48718602 | 0.49 |
ENSMUST00000127537.1
ENSMUST00000155017.2 |
RP24-315D19.2
|
GTPase IMAP family member 1 |
| chr1_+_172499948 | 0.49 |
ENSMUST00000111230.1
|
Tagln2
|
transgelin 2 |
| chr1_+_132880273 | 0.48 |
ENSMUST00000027706.3
|
Lrrn2
|
leucine rich repeat protein 2, neuronal |
| chr4_-_149698552 | 0.48 |
ENSMUST00000134534.1
ENSMUST00000146612.1 ENSMUST00000105688.3 |
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide |
| chr2_-_24935148 | 0.46 |
ENSMUST00000102935.3
ENSMUST00000133934.1 ENSMUST00000028349.7 |
Arrdc1
|
arrestin domain containing 1 |
| chr19_-_47919269 | 0.46 |
ENSMUST00000095998.5
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chrX_-_7671341 | 0.46 |
ENSMUST00000033486.5
|
Plp2
|
proteolipid protein 2 |
| chr1_+_179803376 | 0.46 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chr17_+_27556613 | 0.46 |
ENSMUST00000117600.1
ENSMUST00000114888.3 |
Hmga1
|
high mobility group AT-hook 1 |
| chr10_+_17796256 | 0.45 |
ENSMUST00000037964.6
|
Txlnb
|
taxilin beta |
| chr11_+_78322965 | 0.45 |
ENSMUST00000017534.8
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr4_+_138262189 | 0.45 |
ENSMUST00000030539.3
|
Kif17
|
kinesin family member 17 |
| chr8_+_94179089 | 0.45 |
ENSMUST00000034215.6
|
Mt1
|
metallothionein 1 |
| chr10_-_5805412 | 0.45 |
ENSMUST00000019907.7
|
Fbxo5
|
F-box protein 5 |
| chr5_+_108065742 | 0.45 |
ENSMUST00000081567.4
ENSMUST00000170319.1 ENSMUST00000112626.1 |
Mtf2
|
metal response element binding transcription factor 2 |
| chr11_+_121421388 | 0.45 |
ENSMUST00000038096.7
|
Fn3krp
|
fructosamine 3 kinase related protein |
| chr1_-_193370225 | 0.44 |
ENSMUST00000169907.1
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma |
| chr19_+_59260878 | 0.44 |
ENSMUST00000026084.3
|
Slc18a2
|
solute carrier family 18 (vesicular monoamine), member 2 |
| chr6_-_129533267 | 0.44 |
ENSMUST00000181594.1
|
1700101I11Rik
|
RIKEN cDNA 1700101I11 gene |
| chr9_+_54699548 | 0.44 |
ENSMUST00000070070.7
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4 |
| chr11_+_50377719 | 0.43 |
ENSMUST00000069304.7
ENSMUST00000077817.7 |
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr7_+_19024387 | 0.43 |
ENSMUST00000153976.1
|
Sympk
|
symplekin |
| chr9_-_67043709 | 0.43 |
ENSMUST00000113689.1
ENSMUST00000113684.1 |
Tpm1
|
tropomyosin 1, alpha |
| chr14_+_56668242 | 0.43 |
ENSMUST00000116468.1
|
Mphosph8
|
M-phase phosphoprotein 8 |
| chr16_-_20426322 | 0.43 |
ENSMUST00000115547.2
ENSMUST00000096199.4 |
Abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr17_+_27556668 | 0.42 |
ENSMUST00000117254.1
ENSMUST00000118570.1 |
Hmga1
|
high mobility group AT-hook 1 |
| chr7_-_25615874 | 0.42 |
ENSMUST00000098663.1
|
Gm7092
|
predicted gene 7092 |
| chr14_+_59625281 | 0.42 |
ENSMUST00000053949.5
|
Shisa2
|
shisa homolog 2 (Xenopus laevis) |
| chr11_+_3332426 | 0.42 |
ENSMUST00000136474.1
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_103914560 | 0.42 |
ENSMUST00000106806.1
|
Rsbn1
|
rosbin, round spermatid basic protein 1 |
| chr4_+_134343466 | 0.42 |
ENSMUST00000105872.1
|
Slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr4_-_32950813 | 0.42 |
ENSMUST00000084750.1
ENSMUST00000084748.2 |
Ankrd6
|
ankyrin repeat domain 6 |
| chr7_-_98178254 | 0.41 |
ENSMUST00000040971.7
|
Capn5
|
calpain 5 |
| chr9_-_67043832 | 0.41 |
ENSMUST00000113686.1
|
Tpm1
|
tropomyosin 1, alpha |
| chr4_+_11191726 | 0.40 |
ENSMUST00000029866.9
ENSMUST00000108324.3 |
Ccne2
|
cyclin E2 |
| chr4_-_149698698 | 0.40 |
ENSMUST00000038859.7
ENSMUST00000105690.2 |
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide |
| chr12_+_33147754 | 0.40 |
ENSMUST00000146040.1
ENSMUST00000125192.1 |
Atxn7l1
|
ataxin 7-like 1 |
| chr2_-_140170528 | 0.40 |
ENSMUST00000046030.7
|
Esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr8_-_123515455 | 0.39 |
ENSMUST00000176286.1
ENSMUST00000169210.1 ENSMUST00000074879.5 ENSMUST00000066198.7 ENSMUST00000176155.1 |
Dbndd1
|
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr4_+_47353283 | 0.39 |
ENSMUST00000044234.7
|
Tgfbr1
|
transforming growth factor, beta receptor I |
| chr16_+_20589471 | 0.39 |
ENSMUST00000100074.3
ENSMUST00000096197.4 |
Vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr6_+_36388055 | 0.38 |
ENSMUST00000172278.1
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr5_+_123394782 | 0.38 |
ENSMUST00000111596.1
ENSMUST00000068237.5 |
Mlxip
|
MLX interacting protein |
| chr7_-_126704179 | 0.38 |
ENSMUST00000106364.1
|
Coro1a
|
coronin, actin binding protein 1A |
| chr10_+_86021961 | 0.38 |
ENSMUST00000130320.1
|
Fbxo7
|
F-box protein 7 |
| chr7_+_4925802 | 0.38 |
ENSMUST00000057612.7
|
Ssc5d
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chr15_-_97831460 | 0.38 |
ENSMUST00000079838.7
ENSMUST00000118294.1 |
Hdac7
|
histone deacetylase 7 |
| chr15_+_102459193 | 0.38 |
ENSMUST00000164957.1
ENSMUST00000171245.1 |
Prr13
|
proline rich 13 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.6 | 4.5 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.4 | 1.7 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.4 | 2.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.4 | 3.5 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.4 | 2.6 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.3 | 0.3 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.3 | 1.8 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.3 | 2.4 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.3 | 1.7 | GO:0033026 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.3 | 1.4 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.3 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 1.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 1.0 | GO:2001245 | negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 | 0.7 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 1.2 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 0.7 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.2 | 1.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.2 | 0.6 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.2 | 0.8 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.2 | 0.5 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.2 | 0.7 | GO:0051316 | attachment of spindle microtubules to kinetochore involved in meiotic chromosome segregation(GO:0051316) |
| 0.2 | 0.9 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.2 | 1.4 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 0.8 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.2 | 0.7 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.2 | 1.0 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.6 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 1.9 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.2 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.5 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.2 | 0.6 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.2 | 0.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 0.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 0.5 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.1 | 4.3 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.7 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.3 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.1 | 2.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 | 0.8 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:1903726 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.4 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.1 | 0.5 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.1 | 0.5 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.6 | GO:1904925 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 5.7 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 0.6 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 | 2.3 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.2 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.3 | GO:2001200 | positive regulation of dendritic cell differentiation(GO:2001200) |
| 0.1 | 0.6 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 1.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.1 | 0.8 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.5 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.1 | 2.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.3 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.3 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.3 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.4 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) |
| 0.1 | 0.5 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.4 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 1.3 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.1 | 0.3 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.3 | GO:0021634 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.1 | 0.7 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 0.9 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.2 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.5 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.1 | 0.4 | GO:0061502 | uropod organization(GO:0032796) early endosome to recycling endosome transport(GO:0061502) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 1.6 | GO:0046852 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.1 | 0.2 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.1 | 0.2 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.2 | GO:0046726 | regulation by virus of viral protein levels in host cell(GO:0046719) positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.1 | 0.4 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.0 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 2.8 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.1 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.9 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.2 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 0.1 | 0.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.6 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:2000812 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0051081 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.3 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.3 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.2 | GO:0048690 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 1.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0001844 | protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:0001844) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.5 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.1 | GO:0038091 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 1.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.6 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.2 | GO:0051790 | acetate metabolic process(GO:0006083) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.6 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.8 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.5 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.3 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.5 | GO:1901739 | regulation of myoblast fusion(GO:1901739) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.0 | 0.3 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.4 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.3 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.2 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.1 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.5 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.8 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.9 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.2 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.0 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.0 | 0.6 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.3 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.7 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.0 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.4 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.5 | 2.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 1.7 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.2 | 0.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.7 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.4 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 1.4 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.6 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.4 | GO:0099524 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 1.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.6 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.6 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.7 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.8 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 1.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.9 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 0.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.6 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 4.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.9 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0044300 | juxtaparanode region of axon(GO:0044224) cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) RISC-loading complex(GO:0070578) |
| 0.0 | 0.1 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.0 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.5 | 2.2 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.5 | 1.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.4 | 1.2 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.3 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 1.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.3 | 1.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 1.3 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 1.0 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 1.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 2.8 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.2 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.2 | 1.8 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.6 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 0.7 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 2.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 1.4 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.0 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.6 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.4 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.4 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.4 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.4 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 2.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.5 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 2.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.5 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.6 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 4.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) Krueppel-associated box domain binding(GO:0035851) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.2 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 1.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.9 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.1 | GO:0051381 | histamine binding(GO:0051381) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 2.1 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 1.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.1 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 3.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.9 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 3.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 2.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 1.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 4.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.7 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 1.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 2.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.0 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.0 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE EXCISION REPAIR | Genes involved in Nucleotide Excision Repair |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |