Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tbx19
Z-value: 2.31
Transcription factors associated with Tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx19
|
ENSMUSG00000026572.5 | T-box 19 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx19 | mm10_v2_chr1_-_165160773_165160781 | -0.70 | 2.1e-06 | Click! |
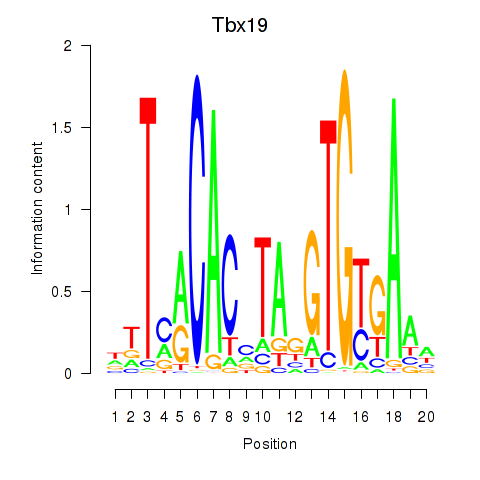
Activity profile of Tbx19 motif
Sorted Z-values of Tbx19 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60499332 | 17.20 |
ENSMUST00000135953.1
|
Mup1
|
major urinary protein 1 |
| chr3_+_138415484 | 16.98 |
ENSMUST00000161312.1
ENSMUST00000013458.8 |
Adh4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr19_-_40187277 | 10.33 |
ENSMUST00000051846.6
|
Cyp2c70
|
cytochrome P450, family 2, subfamily c, polypeptide 70 |
| chr17_+_79626669 | 8.47 |
ENSMUST00000086570.1
|
4921513D11Rik
|
RIKEN cDNA 4921513D11 gene |
| chr6_-_119467210 | 7.79 |
ENSMUST00000118120.1
|
Wnt5b
|
wingless-related MMTV integration site 5B |
| chr17_+_36942910 | 7.00 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr3_+_94693556 | 6.71 |
ENSMUST00000090848.3
ENSMUST00000173981.1 ENSMUST00000173849.1 ENSMUST00000174223.1 |
Selenbp2
|
selenium binding protein 2 |
| chr19_+_41029275 | 6.63 |
ENSMUST00000051806.4
ENSMUST00000112200.1 |
Dntt
|
deoxynucleotidyltransferase, terminal |
| chr14_-_66124482 | 6.28 |
ENSMUST00000070515.1
|
Ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr19_+_12633303 | 6.08 |
ENSMUST00000044976.5
|
Glyat
|
glycine-N-acyltransferase |
| chr10_+_88459569 | 6.00 |
ENSMUST00000020252.3
ENSMUST00000125612.1 |
Sycp3
|
synaptonemal complex protein 3 |
| chr10_-_24927444 | 5.60 |
ENSMUST00000020161.8
|
Arg1
|
arginase, liver |
| chr16_+_48842552 | 5.46 |
ENSMUST00000023329.4
|
Retnla
|
resistin like alpha |
| chr9_-_119157055 | 5.10 |
ENSMUST00000010795.4
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr14_+_66140919 | 4.98 |
ENSMUST00000022620.9
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr4_+_155562348 | 4.53 |
ENSMUST00000030939.7
|
Nadk
|
NAD kinase |
| chr11_+_101468164 | 4.41 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr17_+_26113286 | 3.80 |
ENSMUST00000025010.7
|
Tmem8
|
transmembrane protein 8 (five membrane-spanning domains) |
| chr1_-_72212249 | 3.76 |
ENSMUST00000048860.7
|
Mreg
|
melanoregulin |
| chr11_-_46698962 | 3.73 |
ENSMUST00000109225.2
|
Timd2
|
T cell immunoglobulin and mucin domain containing 2 |
| chr6_+_90465287 | 3.73 |
ENSMUST00000113530.1
|
Klf15
|
Kruppel-like factor 15 |
| chr8_-_41133697 | 3.66 |
ENSMUST00000155055.1
ENSMUST00000059115.6 ENSMUST00000145860.1 |
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr6_+_129180613 | 3.58 |
ENSMUST00000032260.5
|
Clec2d
|
C-type lectin domain family 2, member d |
| chr10_-_93540016 | 3.29 |
ENSMUST00000016034.2
|
Amdhd1
|
amidohydrolase domain containing 1 |
| chr1_+_171419027 | 3.17 |
ENSMUST00000171362.1
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chrM_+_9870 | 3.11 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L |
| chrM_+_10167 | 3.07 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr19_+_8837445 | 2.91 |
ENSMUST00000171649.1
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human) |
| chr13_+_58806564 | 2.81 |
ENSMUST00000109838.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr1_+_185332143 | 2.78 |
ENSMUST00000027916.6
ENSMUST00000151769.1 ENSMUST00000110965.1 |
Bpnt1
|
bisphosphate 3'-nucleotidase 1 |
| chr12_-_80968075 | 2.75 |
ENSMUST00000095572.4
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr7_+_130936172 | 2.71 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr2_-_113758638 | 2.64 |
ENSMUST00000099575.3
|
Grem1
|
gremlin 1 |
| chr11_-_101171302 | 2.62 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr9_-_103230262 | 2.53 |
ENSMUST00000165296.1
ENSMUST00000112645.1 ENSMUST00000166836.1 |
Trf
Gm20425
|
transferrin predicted gene 20425 |
| chr1_-_24612700 | 2.53 |
ENSMUST00000088336.1
|
Gm10222
|
predicted gene 10222 |
| chr5_-_115484297 | 2.43 |
ENSMUST00000112067.1
|
Sirt4
|
sirtuin 4 |
| chr14_-_55585250 | 2.36 |
ENSMUST00000022828.8
|
Emc9
|
ER membrane protein complex subunit 9 |
| chr17_+_34670535 | 2.36 |
ENSMUST00000168533.1
ENSMUST00000087399.4 |
Tnxb
|
tenascin XB |
| chr5_-_38491948 | 2.36 |
ENSMUST00000129099.1
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr5_+_34525797 | 2.27 |
ENSMUST00000125817.1
ENSMUST00000067638.7 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr4_-_12087912 | 2.24 |
ENSMUST00000050686.3
|
Tmem67
|
transmembrane protein 67 |
| chr17_+_6978860 | 2.24 |
ENSMUST00000089119.5
ENSMUST00000179728.1 |
Rnaset2b
|
ribonuclease T2B |
| chr5_-_138996087 | 2.22 |
ENSMUST00000110897.1
|
Pdgfa
|
platelet derived growth factor, alpha |
| chr3_-_88254706 | 2.18 |
ENSMUST00000171887.1
|
Rhbg
|
Rhesus blood group-associated B glycoprotein |
| chr9_+_108569885 | 2.13 |
ENSMUST00000019183.8
|
Dalrd3
|
DALR anticodon binding domain containing 3 |
| chr14_+_65666394 | 2.08 |
ENSMUST00000022610.8
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chr2_-_148046896 | 2.03 |
ENSMUST00000172928.1
ENSMUST00000047315.3 |
Foxa2
|
forkhead box A2 |
| chr2_-_112368021 | 2.01 |
ENSMUST00000028551.3
|
Emc4
|
ER membrane protein complex subunit 4 |
| chr7_+_12927410 | 2.01 |
ENSMUST00000045870.4
|
2310014L17Rik
|
RIKEN cDNA 2310014L17 gene |
| chr11_-_60046477 | 1.99 |
ENSMUST00000000310.7
ENSMUST00000102693.2 ENSMUST00000148512.1 |
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr7_+_105404568 | 1.98 |
ENSMUST00000033187.4
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr15_+_31602106 | 1.95 |
ENSMUST00000042702.6
|
Fam173b
|
family with sequence similarity 173, member B |
| chr16_-_3908639 | 1.94 |
ENSMUST00000115859.1
|
1700037C18Rik
|
RIKEN cDNA 1700037C18 gene |
| chrX_+_133908441 | 1.92 |
ENSMUST00000113304.1
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr7_-_140955960 | 1.91 |
ENSMUST00000081649.8
|
Ifitm2
|
interferon induced transmembrane protein 2 |
| chr13_-_52929640 | 1.81 |
ENSMUST00000120535.1
ENSMUST00000119311.1 ENSMUST00000021913.9 ENSMUST00000110031.3 |
Auh
|
AU RNA binding protein/enoyl-coenzyme A hydratase |
| chr7_+_143830077 | 1.81 |
ENSMUST00000141916.1
|
Dhcr7
|
7-dehydrocholesterol reductase |
| chr3_-_30509462 | 1.78 |
ENSMUST00000173899.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chrX_+_140664908 | 1.76 |
ENSMUST00000112990.1
ENSMUST00000112988.1 |
Mid2
|
midline 2 |
| chr1_+_59256906 | 1.76 |
ENSMUST00000160662.1
ENSMUST00000114248.2 |
Cdk15
|
cyclin-dependent kinase 15 |
| chr17_+_25773769 | 1.75 |
ENSMUST00000134108.1
ENSMUST00000002350.4 |
Narfl
|
nuclear prelamin A recognition factor-like |
| chr19_+_3958803 | 1.69 |
ENSMUST00000179433.1
|
1700055N04Rik
|
RIKEN cDNA 1700055N04 gene |
| chr19_-_41206774 | 1.69 |
ENSMUST00000025986.7
ENSMUST00000169941.1 |
Tll2
|
tolloid-like 2 |
| chr19_-_11818806 | 1.68 |
ENSMUST00000075304.6
|
Stx3
|
syntaxin 3 |
| chr7_+_143830204 | 1.67 |
ENSMUST00000144034.1
ENSMUST00000143338.1 |
Dhcr7
|
7-dehydrocholesterol reductase |
| chr11_+_72301613 | 1.66 |
ENSMUST00000151440.1
ENSMUST00000146233.1 ENSMUST00000140842.2 |
Xaf1
|
XIAP associated factor 1 |
| chr7_+_48959089 | 1.63 |
ENSMUST00000183659.1
|
Nav2
|
neuron navigator 2 |
| chr17_+_56628118 | 1.60 |
ENSMUST00000112979.2
|
Catsperd
|
catsper channel auxiliary subunit delta |
| chr13_-_56482246 | 1.49 |
ENSMUST00000022019.3
|
Il9
|
interleukin 9 |
| chr14_+_65666430 | 1.47 |
ENSMUST00000069226.6
|
Scara5
|
scavenger receptor class A, member 5 (putative) |
| chr1_-_59094885 | 1.47 |
ENSMUST00000097080.3
|
Als2cr11
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 (human) |
| chrX_+_73787002 | 1.44 |
ENSMUST00000166518.1
|
Ssr4
|
signal sequence receptor, delta |
| chr7_-_80947765 | 1.44 |
ENSMUST00000026818.5
ENSMUST00000117383.1 ENSMUST00000119980.1 |
Sec11a
|
SEC11 homolog A (S. cerevisiae) |
| chr4_+_122836236 | 1.43 |
ENSMUST00000030412.4
ENSMUST00000121870.1 ENSMUST00000097902.4 |
Ppt1
|
palmitoyl-protein thioesterase 1 |
| chr18_-_82406777 | 1.40 |
ENSMUST00000065224.6
|
Galr1
|
galanin receptor 1 |
| chr1_+_33719863 | 1.39 |
ENSMUST00000088287.3
|
Rab23
|
RAB23, member RAS oncogene family |
| chr19_-_10920629 | 1.38 |
ENSMUST00000025641.1
|
Zp1
|
zona pellucida glycoprotein 1 |
| chrX_+_100428906 | 1.38 |
ENSMUST00000060241.2
|
Otud6a
|
OTU domain containing 6A |
| chr19_+_34290653 | 1.33 |
ENSMUST00000025691.5
ENSMUST00000112472.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr13_-_67332525 | 1.30 |
ENSMUST00000168892.1
ENSMUST00000109735.2 |
Zfp595
|
zinc finger protein 595 |
| chr10_-_86011833 | 1.27 |
ENSMUST00000105304.1
ENSMUST00000061699.5 |
Bpifc
|
BPI fold containing family C |
| chr1_-_39805311 | 1.27 |
ENSMUST00000171319.2
|
Gm3646
|
predicted gene 3646 |
| chr7_+_28741968 | 1.26 |
ENSMUST00000094632.4
|
Sars2
|
seryl-aminoacyl-tRNA synthetase 2 |
| chr11_+_69326252 | 1.25 |
ENSMUST00000018614.2
|
Kcnab3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr16_-_92466081 | 1.19 |
ENSMUST00000060005.8
|
Rcan1
|
regulator of calcineurin 1 |
| chr6_+_124808885 | 1.17 |
ENSMUST00000143040.1
ENSMUST00000052727.4 ENSMUST00000130160.1 |
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chrX_+_32894625 | 1.16 |
ENSMUST00000179069.1
|
Gm21657
|
predicted gene, 21657 |
| chr9_-_16378231 | 1.15 |
ENSMUST00000082170.5
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila) |
| chr9_+_18404418 | 1.15 |
ENSMUST00000164441.1
ENSMUST00000169398.1 |
Ubtfl1
|
upstream binding transcription factor, RNA polymerase I-like 1 |
| chr2_+_176236860 | 1.13 |
ENSMUST00000166464.1
|
2210418O10Rik
|
RIKEN cDNA 2210418O10 gene |
| chr4_+_143615003 | 1.13 |
ENSMUST00000105773.1
|
Gm13083
|
predicted gene 13083 |
| chr9_+_58582397 | 1.12 |
ENSMUST00000176557.1
ENSMUST00000114121.4 ENSMUST00000177064.1 |
Nptn
|
neuroplastin |
| chr4_-_84546284 | 1.12 |
ENSMUST00000177040.1
|
Bnc2
|
basonuclin 2 |
| chr13_+_4771649 | 1.12 |
ENSMUST00000065956.3
|
Gm5444
|
predicted gene 5444 |
| chr6_+_17749170 | 1.12 |
ENSMUST00000053148.7
ENSMUST00000115417.3 |
St7
|
suppression of tumorigenicity 7 |
| chrX_-_34161499 | 1.11 |
ENSMUST00000178219.1
|
Gm21681
|
predicted gene, 21681 |
| chrX_+_31117674 | 1.10 |
ENSMUST00000179532.1
|
Gm21447
|
predicted gene, 21447 |
| chr14_-_63543931 | 1.07 |
ENSMUST00000058679.5
|
Mtmr9
|
myotubularin related protein 9 |
| chr9_-_25151772 | 1.06 |
ENSMUST00000008573.7
|
Herpud2
|
HERPUD family member 2 |
| chrX_+_32047829 | 1.06 |
ENSMUST00000178747.1
|
Gm21645
|
predicted gene, 21645 |
| chrX_-_33956540 | 1.06 |
ENSMUST00000179466.1
|
Gm2964
|
predicted gene 2964 |
| chr8_+_105225145 | 1.06 |
ENSMUST00000034361.3
|
D230025D16Rik
|
RIKEN cDNA D230025D16 gene |
| chrX_+_133908418 | 1.02 |
ENSMUST00000033606.8
ENSMUST00000113303.1 ENSMUST00000165805.1 |
Srpx2
|
sushi-repeat-containing protein, X-linked 2 |
| chr2_-_175067763 | 1.00 |
ENSMUST00000072895.3
ENSMUST00000109066.1 |
Gm14393
|
predicted gene 14393 |
| chr9_-_57606234 | 0.95 |
ENSMUST00000045068.8
|
Cplx3
|
complexin 3 |
| chr7_-_110844350 | 0.93 |
ENSMUST00000177462.1
ENSMUST00000176746.1 ENSMUST00000177236.1 |
Rnf141
|
ring finger protein 141 |
| chr13_+_3538075 | 0.92 |
ENSMUST00000059515.6
|
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2 |
| chr14_+_53845234 | 0.89 |
ENSMUST00000103674.4
|
Trav19
|
T cell receptor alpha variable 19 |
| chrX_-_3443690 | 0.87 |
ENSMUST00000105020.1
|
Gm14346
|
predicted gene 14346 |
| chr2_-_181135220 | 0.85 |
ENSMUST00000016491.7
|
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2 |
| chr17_+_46646225 | 0.85 |
ENSMUST00000002844.7
ENSMUST00000113429.1 ENSMUST00000113430.1 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr16_-_45844303 | 0.84 |
ENSMUST00000036355.6
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr14_-_50870557 | 0.83 |
ENSMUST00000006444.7
|
Tep1
|
telomerase associated protein 1 |
| chr13_-_67332498 | 0.81 |
ENSMUST00000171466.1
|
Zfp595
|
zinc finger protein 595 |
| chrX_+_3700233 | 0.81 |
ENSMUST00000105019.2
|
Gm14345
|
predicted gene 14345 |
| chrX_-_33577346 | 0.81 |
ENSMUST00000105119.3
|
Gm2913
|
predicted gene 2913 |
| chrX_+_32560055 | 0.81 |
ENSMUST00000178070.1
|
Gmcl1l
|
germ cell-less homolog 1 (Drosophila)-like |
| chrX_-_3752885 | 0.81 |
ENSMUST00000178621.1
|
Gm14351
|
predicted gene 14351 |
| chrX_+_32278394 | 0.81 |
ENSMUST00000179991.1
|
Gm2799
|
predicted gene 2799 |
| chr15_-_98379197 | 0.81 |
ENSMUST00000057386.5
|
Olfr283
|
olfactory receptor 283 |
| chrX_+_4196576 | 0.79 |
ENSMUST00000105015.2
|
Gm14347
|
predicted gene 14347 |
| chrX_-_31383918 | 0.79 |
ENSMUST00000178444.1
|
Gm2777
|
predicted gene 2777 |
| chrX_+_4370636 | 0.79 |
ENSMUST00000105014.1
|
Gm10922
|
predicted gene 10922 |
| chr2_-_177578199 | 0.78 |
ENSMUST00000108945.1
ENSMUST00000108943.1 |
Gm14406
|
predicted gene 14406 |
| chr2_+_175010241 | 0.78 |
ENSMUST00000109069.1
ENSMUST00000109070.2 |
Gm14444
|
predicted gene 14444 |
| chrX_-_3957002 | 0.78 |
ENSMUST00000105017.3
|
Gm3701
|
predicted gene 3701 |
| chrX_+_33313338 | 0.78 |
ENSMUST00000180039.1
|
Gm2863
|
predicted gene 2863 |
| chrX_+_32973897 | 0.78 |
ENSMUST00000179538.1
|
Gm2825
|
predicted gene 2825 |
| chrX_-_4291245 | 0.78 |
ENSMUST00000179325.1
|
Gm10921
|
predicted gene 10921 |
| chr5_-_121191365 | 0.78 |
ENSMUST00000100770.2
ENSMUST00000054547.7 |
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr15_-_36140393 | 0.76 |
ENSMUST00000172831.1
|
Rgs22
|
regulator of G-protein signalling 22 |
| chr1_+_127868773 | 0.75 |
ENSMUST00000037649.5
|
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chrX_+_32726137 | 0.75 |
ENSMUST00000179585.1
|
Gm21951
|
predicted gene, 21951 |
| chr6_+_86195214 | 0.74 |
ENSMUST00000032066.9
|
Tgfa
|
transforming growth factor alpha |
| chrX_+_33657172 | 0.74 |
ENSMUST00000177912.1
|
Gm2927
|
predicted gene 2927 |
| chr16_-_44746278 | 0.74 |
ENSMUST00000161436.1
|
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chrX_+_14211148 | 0.72 |
ENSMUST00000079952.2
|
Gm5382
|
predicted gene 5382 |
| chr3_-_88372740 | 0.72 |
ENSMUST00000107543.1
ENSMUST00000107542.1 |
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr2_+_153875045 | 0.72 |
ENSMUST00000028983.2
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr6_+_41302265 | 0.69 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chrX_-_33702349 | 0.68 |
ENSMUST00000105117.2
|
Gm2933
|
predicted gene 2933 |
| chr16_-_44746337 | 0.67 |
ENSMUST00000023348.4
ENSMUST00000162512.1 |
Gtpbp8
|
GTP-binding protein 8 (putative) |
| chr14_-_56120623 | 0.67 |
ENSMUST00000089549.6
|
Gzme
|
granzyme E |
| chr8_+_129072644 | 0.67 |
ENSMUST00000148234.1
|
1700008F21Rik
|
RIKEN cDNA 1700008F21 gene |
| chr9_-_6266544 | 0.65 |
ENSMUST00000051706.4
|
Ddi1
|
DDI1, DNA-damage inducible 1, homolog 1 (S. cerevisiae) |
| chr11_-_69920892 | 0.63 |
ENSMUST00000152589.1
ENSMUST00000108612.1 ENSMUST00000108611.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_15712303 | 0.61 |
ENSMUST00000170146.1
|
Kcnb2
|
potassium voltage gated channel, Shab-related subfamily, member 2 |
| chr1_-_164935522 | 0.61 |
ENSMUST00000027860.7
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr15_-_100403243 | 0.60 |
ENSMUST00000124324.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr18_+_37484955 | 0.58 |
ENSMUST00000053856.4
|
Pcdhb17
|
protocadherin beta 17 |
| chr15_-_101869695 | 0.57 |
ENSMUST00000087996.5
|
Krt77
|
keratin 77 |
| chr9_-_57467985 | 0.55 |
ENSMUST00000046587.6
|
Scamp5
|
secretory carrier membrane protein 5 |
| chr1_+_183388981 | 0.55 |
ENSMUST00000097043.5
|
Taf1a
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, A |
| chr7_-_134364859 | 0.54 |
ENSMUST00000172947.1
|
D7Ertd443e
|
DNA segment, Chr 7, ERATO Doi 443, expressed |
| chr17_+_13234871 | 0.54 |
ENSMUST00000059824.6
|
Smok2b
|
sperm motility kinase 2B |
| chr5_-_143527977 | 0.52 |
ENSMUST00000100489.3
ENSMUST00000080537.7 |
Rac1
|
RAS-related C3 botulinum substrate 1 |
| chr2_+_177080256 | 0.50 |
ENSMUST00000134614.1
ENSMUST00000108968.1 |
Gm14401
|
predicted gene 14401 |
| chr5_+_96173940 | 0.49 |
ENSMUST00000048361.8
|
Gm2840
|
predicted gene 2840 |
| chr6_+_42286709 | 0.49 |
ENSMUST00000163936.1
|
Clcn1
|
chloride channel 1 |
| chr7_-_132599637 | 0.49 |
ENSMUST00000054562.3
|
Nkx1-2
|
NK1 transcription factor related, locus 2 (Drosophila) |
| chr11_+_49340534 | 0.48 |
ENSMUST00000062719.1
|
Olfr1390
|
olfactory receptor 1390 |
| chr2_+_181991226 | 0.45 |
ENSMUST00000071760.6
|
Gm14496
|
predicted gene 14496 |
| chr19_+_59219648 | 0.41 |
ENSMUST00000065204.6
|
Kcnk18
|
potassium channel, subfamily K, member 18 |
| chr11_-_115322025 | 0.39 |
ENSMUST00000103037.4
|
Ush1g
|
Usher syndrome 1G |
| chr1_+_159232299 | 0.36 |
ENSMUST00000076894.5
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr7_+_22412231 | 0.34 |
ENSMUST00000177774.1
|
Vmn1r148
|
vomeronasal 1 receptor 148 |
| chr4_-_144274027 | 0.33 |
ENSMUST00000105752.3
|
Pramel5
|
preferentially expressed antigen in melanoma like 5 |
| chr5_-_129704259 | 0.33 |
ENSMUST00000042266.6
|
Sept14
|
septin 14 |
| chr1_-_78488846 | 0.31 |
ENSMUST00000068333.7
ENSMUST00000170217.1 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chrX_+_11305655 | 0.30 |
ENSMUST00000178806.1
|
Gm14477
|
predicted gene 14477 |
| chr12_-_58269162 | 0.28 |
ENSMUST00000062254.2
|
Clec14a
|
C-type lectin domain family 14, member a |
| chr12_-_98259416 | 0.26 |
ENSMUST00000021390.7
|
Galc
|
galactosylceramidase |
| chr13_-_74376566 | 0.24 |
ENSMUST00000091481.2
|
Zfp72
|
zinc finger protein 72 |
| chr9_-_124440899 | 0.24 |
ENSMUST00000180233.1
|
Gm20783
|
predicted gene, 20783 |
| chr3_-_97610156 | 0.23 |
ENSMUST00000029730.4
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr3_-_153944632 | 0.22 |
ENSMUST00000072697.6
|
Acadm
|
acyl-Coenzyme A dehydrogenase, medium chain |
| chr6_-_54593139 | 0.22 |
ENSMUST00000046520.6
|
Fkbp14
|
FK506 binding protein 14 |
| chr17_-_33216351 | 0.21 |
ENSMUST00000112162.3
|
Gm4461
|
predicted gene 4461 |
| chr8_-_4275886 | 0.18 |
ENSMUST00000003029.7
|
Timm44
|
translocase of inner mitochondrial membrane 44 |
| chr2_-_177324307 | 0.18 |
ENSMUST00000108959.2
|
Gm14412
|
predicted gene 14412 |
| chr15_+_31276491 | 0.17 |
ENSMUST00000068987.5
|
Fam136b-ps
|
family with sequence similarity 136, member B, pseudogene |
| chr13_-_21453714 | 0.15 |
ENSMUST00000032820.7
ENSMUST00000110485.1 |
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr4_+_43384332 | 0.15 |
ENSMUST00000136360.1
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr7_+_17713233 | 0.13 |
ENSMUST00000081907.6
|
Ceacam5
|
carcinoembryonic antigen-related cell adhesion molecule 5 |
| chr1_+_156838915 | 0.10 |
ENSMUST00000111720.1
|
Angptl1
|
angiopoietin-like 1 |
| chr9_+_58582240 | 0.09 |
ENSMUST00000177292.1
ENSMUST00000085651.5 |
Nptn
|
neuroplastin |
| chr2_+_88817173 | 0.08 |
ENSMUST00000072057.1
|
Olfr1202
|
olfactory receptor 1202 |
| chr7_-_5014645 | 0.06 |
ENSMUST00000165320.1
|
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr14_-_52036143 | 0.05 |
ENSMUST00000052560.4
|
Olfr221
|
olfactory receptor 221 |
| chr1_-_57406443 | 0.01 |
ENSMUST00000160837.1
ENSMUST00000161780.1 |
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr5_-_45856496 | 0.00 |
ENSMUST00000087164.3
ENSMUST00000121573.1 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr9_+_15520830 | 0.00 |
ENSMUST00000178999.1
|
Smco4
|
single-pass membrane protein with coiled-coil domains 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx19
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 17.0 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 2.1 | 6.3 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) olefin metabolic process(GO:1900673) |
| 1.6 | 17.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 1.5 | 6.0 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) female meiosis sister chromatid cohesion(GO:0007066) |
| 1.4 | 5.6 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 1.2 | 3.5 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.9 | 2.6 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.8 | 4.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 2.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.7 | 2.0 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.7 | 3.3 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.5 | 2.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.5 | 2.4 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.5 | 1.4 | GO:1990168 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.5 | 2.7 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.4 | 1.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.4 | 3.6 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 4.3 | GO:0097286 | iron ion import(GO:0097286) |
| 0.4 | 2.8 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.3 | 1.6 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 2.5 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.3 | 2.7 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 | 8.8 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.3 | 0.8 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.3 | 2.0 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.2 | 1.7 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.2 | 1.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 4.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.2 | 1.4 | GO:0098734 | protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 3.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.2 | 2.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.7 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 1.7 | GO:0098881 | synaptic vesicle docking(GO:0016081) exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.6 | GO:2000563 | immunoglobulin production in mucosal tissue(GO:0002426) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) positive regulation of thymocyte migration(GO:2000412) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.2 | 2.9 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 6.6 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 1.3 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.2 | 2.2 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 3.1 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 3.7 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.2 | 1.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 3.8 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 5.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 2.4 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 9.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 2.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 1.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 3.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 0.6 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.4 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.8 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 1.9 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.2 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 | 0.5 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.1 | 1.5 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.1 | 1.4 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.1 | 3.1 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 1.1 | GO:0043586 | tongue development(GO:0043586) |
| 0.1 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.6 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.5 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 6.1 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 1.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.1 | GO:0001832 | blastocyst growth(GO:0001832) |
| 0.0 | 0.2 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 1.7 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.5 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 1.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 3.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.8 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 5.1 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 2.7 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 1.7 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 1.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 2.5 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 4.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 1.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 5.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 2.0 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 2.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.2 | 3.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 6.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.1 | 0.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 2.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 6.2 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.1 | 1.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 11.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 6.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 5.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 4.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 3.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 24.9 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 2.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 3.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 22.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 17.2 | GO:0005186 | insulin-activated receptor activity(GO:0005009) pheromone activity(GO:0005186) |
| 5.7 | 17.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 1.5 | 6.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.5 | 7.3 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 1.3 | 6.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 1.2 | 3.5 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.7 | 2.8 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.7 | 2.6 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.6 | 6.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 5.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.5 | 3.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.5 | 2.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 1.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.4 | 1.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.4 | 2.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 2.5 | GO:0072510 | signal recognition particle binding(GO:0005047) ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.3 | 5.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 2.0 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.3 | 5.0 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.3 | 8.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 1.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 7.9 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.2 | 6.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.9 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.2 | 4.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.8 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 0.7 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 2.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 2.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.2 | 7.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 2.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 2.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 1.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 2.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.1 | 2.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 2.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.4 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 0.5 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.2 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.9 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 1.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 2.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 2.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 3.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 2.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 3.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 2.2 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.5 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 2.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 2.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 5.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 7.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 3.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 2.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 2.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 4.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.9 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 3.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 4.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 17.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 5.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 2.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 2.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 2.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 3.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 6.1 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 6.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.4 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 3.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 8.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.8 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |