Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Srf
Z-value: 0.93
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSMUSG00000015605.5 | serum response factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | mm10_v2_chr17_-_46556158_46556188 | 0.36 | 3.0e-02 | Click! |
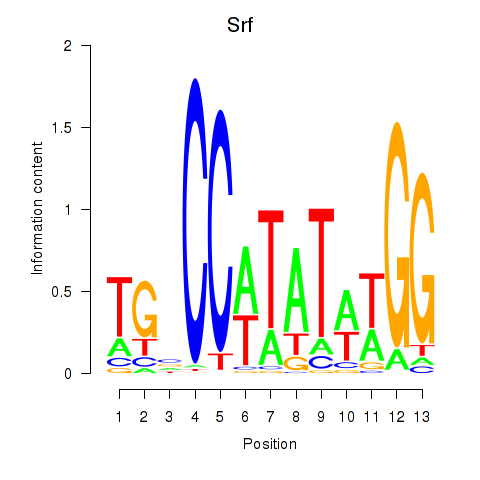
Activity profile of Srf motif
Sorted Z-values of Srf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142661305 | 11.89 |
ENSMUST00000105936.1
|
Igf2
|
insulin-like growth factor 2 |
| chr2_-_114052804 | 9.75 |
ENSMUST00000090269.6
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr8_-_123894736 | 8.13 |
ENSMUST00000034453.4
|
Acta1
|
actin, alpha 1, skeletal muscle |
| chr7_-_142661858 | 6.08 |
ENSMUST00000145896.2
|
Igf2
|
insulin-like growth factor 2 |
| chr4_+_46039202 | 5.27 |
ENSMUST00000156200.1
|
Tmod1
|
tropomodulin 1 |
| chrX_+_101449078 | 4.71 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr7_+_19411086 | 3.72 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr11_-_94973447 | 3.63 |
ENSMUST00000100551.4
ENSMUST00000152042.1 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr8_+_15057646 | 3.43 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr6_-_83536215 | 3.28 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_-_111997204 | 3.19 |
ENSMUST00000074805.5
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr9_+_110763646 | 3.17 |
ENSMUST00000079784.7
|
Myl3
|
myosin, light polypeptide 3 |
| chr4_-_119190005 | 2.82 |
ENSMUST00000138395.1
ENSMUST00000156746.1 |
Ermap
|
erythroblast membrane-associated protein |
| chr2_+_127336152 | 2.78 |
ENSMUST00000028846.6
|
Dusp2
|
dual specificity phosphatase 2 |
| chr14_-_34588654 | 2.76 |
ENSMUST00000022328.6
ENSMUST00000064098.6 ENSMUST00000022327.5 ENSMUST00000022330.7 |
Ldb3
|
LIM domain binding 3 |
| chr4_+_43957401 | 2.69 |
ENSMUST00000030202.7
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr2_+_154548888 | 2.64 |
ENSMUST00000045116.4
ENSMUST00000109709.3 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr10_+_45335751 | 2.58 |
ENSMUST00000095715.3
|
Bves
|
blood vessel epicardial substance |
| chr14_-_54966570 | 2.43 |
ENSMUST00000124930.1
ENSMUST00000134256.1 ENSMUST00000081857.6 ENSMUST00000145322.1 |
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha |
| chr17_-_57194170 | 2.17 |
ENSMUST00000005976.6
|
Tnfsf14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr4_+_43957678 | 2.08 |
ENSMUST00000107855.1
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr2_+_156775409 | 2.00 |
ENSMUST00000088552.6
|
Myl9
|
myosin, light polypeptide 9, regulatory |
| chr2_+_164940742 | 1.85 |
ENSMUST00000137626.1
|
Mmp9
|
matrix metallopeptidase 9 |
| chr5_-_24329556 | 1.65 |
ENSMUST00000115098.2
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr6_-_40951826 | 1.65 |
ENSMUST00000073642.5
|
Gm4744
|
predicted gene 4744 |
| chr4_-_63403330 | 1.64 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chr1_-_43163891 | 1.63 |
ENSMUST00000008280.7
|
Fhl2
|
four and a half LIM domains 2 |
| chr2_-_152415044 | 1.61 |
ENSMUST00000099207.3
|
Zcchc3
|
zinc finger, CCHC domain containing 3 |
| chr14_-_34588607 | 1.57 |
ENSMUST00000090040.4
|
Ldb3
|
LIM domain binding 3 |
| chr11_-_120348513 | 1.44 |
ENSMUST00000071555.6
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr11_-_120348475 | 1.38 |
ENSMUST00000062147.7
ENSMUST00000128055.1 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr3_+_131112785 | 1.38 |
ENSMUST00000098611.3
|
Lef1
|
lymphoid enhancer binding factor 1 |
| chr19_+_53529100 | 1.33 |
ENSMUST00000038287.6
|
Dusp5
|
dual specificity phosphatase 5 |
| chr10_+_4266323 | 1.23 |
ENSMUST00000045730.5
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr5_-_112896350 | 1.18 |
ENSMUST00000086617.4
|
Myo18b
|
myosin XVIIIb |
| chr14_-_19585135 | 1.18 |
ENSMUST00000170694.1
|
Gm2237
|
predicted gene 2237 |
| chr1_+_74409376 | 1.16 |
ENSMUST00000027366.6
|
Vil1
|
villin 1 |
| chr12_-_32953772 | 1.16 |
ENSMUST00000180391.1
ENSMUST00000181670.1 |
4933406C10Rik
|
RIKEN cDNA 4933406C10 gene |
| chr1_-_127677923 | 1.15 |
ENSMUST00000160616.1
|
Tmem163
|
transmembrane protein 163 |
| chr1_-_33757711 | 1.06 |
ENSMUST00000044691.7
|
Bag2
|
BCL2-associated athanogene 2 |
| chr10_+_79988584 | 1.04 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chr10_+_33083476 | 1.03 |
ENSMUST00000095762.4
|
Trdn
|
triadin |
| chr18_+_34861200 | 0.96 |
ENSMUST00000165033.1
|
Egr1
|
early growth response 1 |
| chr4_+_148000722 | 0.94 |
ENSMUST00000103230.4
|
Nppa
|
natriuretic peptide type A |
| chr2_+_150786735 | 0.93 |
ENSMUST00000045441.7
|
Pygb
|
brain glycogen phosphorylase |
| chr19_-_5912771 | 0.92 |
ENSMUST00000118623.1
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr11_-_120348091 | 0.91 |
ENSMUST00000106215.4
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr7_+_128246812 | 0.86 |
ENSMUST00000164710.1
ENSMUST00000070656.5 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr7_+_128246953 | 0.83 |
ENSMUST00000167965.1
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr2_+_125068118 | 0.83 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr11_-_5898771 | 0.82 |
ENSMUST00000102921.3
|
Myl7
|
myosin, light polypeptide 7, regulatory |
| chr4_+_132351768 | 0.82 |
ENSMUST00000172202.1
|
Gm17300
|
predicted gene, 17300 |
| chr4_-_132351636 | 0.76 |
ENSMUST00000105951.1
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr19_-_5912834 | 0.76 |
ENSMUST00000136983.1
|
Dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr11_+_101330605 | 0.73 |
ENSMUST00000103105.3
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr17_+_7925990 | 0.72 |
ENSMUST00000036370.7
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr11_-_99493112 | 0.71 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr1_+_63176818 | 0.71 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr6_-_97179100 | 0.69 |
ENSMUST00000095664.3
|
Tmf1
|
TATA element modulatory factor 1 |
| chr4_+_120615816 | 0.68 |
ENSMUST00000148457.1
|
Gm12860
|
predicted gene 12860 |
| chr13_+_23746734 | 0.66 |
ENSMUST00000099703.2
|
Hist1h2bb
|
histone cluster 1, H2bb |
| chr2_-_91649785 | 0.65 |
ENSMUST00000111333.1
|
Zfp408
|
zinc finger protein 408 |
| chr10_+_67537861 | 0.61 |
ENSMUST00000048289.7
ENSMUST00000105438.2 ENSMUST00000130933.1 ENSMUST00000146986.1 |
Egr2
|
early growth response 2 |
| chr11_-_120731944 | 0.61 |
ENSMUST00000154565.1
ENSMUST00000026148.2 |
Cbr2
|
carbonyl reductase 2 |
| chr4_-_155056784 | 0.58 |
ENSMUST00000131173.2
|
Plch2
|
phospholipase C, eta 2 |
| chr5_+_90367204 | 0.54 |
ENSMUST00000068250.3
|
Gm9958
|
predicted gene 9958 |
| chr8_+_47675362 | 0.52 |
ENSMUST00000098781.2
|
AA386476
|
expressed sequence AA386476 |
| chr3_+_68468162 | 0.52 |
ENSMUST00000182532.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr9_+_53537021 | 0.51 |
ENSMUST00000035850.7
|
Npat
|
nuclear protein in the AT region |
| chr3_-_152266320 | 0.51 |
ENSMUST00000046045.8
|
Nexn
|
nexilin |
| chr11_+_77462325 | 0.51 |
ENSMUST00000102493.1
|
Coro6
|
coronin 6 |
| chr7_+_101896817 | 0.49 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr5_+_24413406 | 0.46 |
ENSMUST00000049346.5
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr7_-_102210120 | 0.45 |
ENSMUST00000070165.5
|
Nup98
|
nucleoporin 98 |
| chr8_+_54550324 | 0.45 |
ENSMUST00000033918.2
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr17_+_5941280 | 0.45 |
ENSMUST00000146009.1
ENSMUST00000115791.2 ENSMUST00000080283.6 |
Synj2
|
synaptojanin 2 |
| chr6_+_87373357 | 0.43 |
ENSMUST00000032128.4
|
Gkn2
|
gastrokine 2 |
| chr9_-_66514567 | 0.42 |
ENSMUST00000056890.8
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr5_+_103754154 | 0.42 |
ENSMUST00000054979.3
|
Aff1
|
AF4/FMR2 family, member 1 |
| chr2_-_60881360 | 0.41 |
ENSMUST00000164147.1
ENSMUST00000112509.1 |
Rbms1
|
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_101896340 | 0.41 |
ENSMUST00000035395.7
ENSMUST00000106973.1 ENSMUST00000144207.1 |
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr16_-_4880284 | 0.38 |
ENSMUST00000037843.6
|
Ubald1
|
UBA-like domain containing 1 |
| chr17_-_73399289 | 0.36 |
ENSMUST00000095208.3
|
Capn13
|
calpain 13 |
| chr19_+_5447692 | 0.36 |
ENSMUST00000025850.5
|
Fosl1
|
fos-like antigen 1 |
| chrX_+_11311934 | 0.36 |
ENSMUST00000178979.1
|
Gm14484
|
predicted gene 14484 |
| chr3_+_106113229 | 0.33 |
ENSMUST00000079132.5
ENSMUST00000139086.1 |
Chia
|
chitinase, acidic |
| chr17_+_23803179 | 0.31 |
ENSMUST00000088621.4
|
Srrm2
|
serine/arginine repetitive matrix 2 |
| chr11_+_101331069 | 0.31 |
ENSMUST00000017316.6
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr12_+_86678685 | 0.31 |
ENSMUST00000021681.3
|
Vash1
|
vasohibin 1 |
| chr3_-_95891929 | 0.29 |
ENSMUST00000171519.1
|
BC028528
|
cDNA sequence BC028528 |
| chr12_-_40199315 | 0.29 |
ENSMUST00000095760.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr2_+_127208358 | 0.28 |
ENSMUST00000103220.3
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr7_-_44997221 | 0.27 |
ENSMUST00000152341.1
|
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr13_+_109260481 | 0.26 |
ENSMUST00000153234.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr5_+_98180866 | 0.25 |
ENSMUST00000112959.1
|
Prdm8
|
PR domain containing 8 |
| chr14_+_53640701 | 0.25 |
ENSMUST00000179267.2
|
Trav14-2
|
T cell receptor alpha variable 14-2 |
| chr3_-_19311269 | 0.24 |
ENSMUST00000099195.3
|
Pde7a
|
phosphodiesterase 7A |
| chr2_-_127521358 | 0.23 |
ENSMUST00000028850.8
ENSMUST00000103215.4 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr2_+_31759932 | 0.21 |
ENSMUST00000028190.6
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr2_-_13271419 | 0.19 |
ENSMUST00000028059.2
|
Rsu1
|
Ras suppressor protein 1 |
| chr12_-_103355967 | 0.19 |
ENSMUST00000021617.7
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr2_-_13271268 | 0.18 |
ENSMUST00000137670.1
ENSMUST00000114791.2 |
Rsu1
|
Ras suppressor protein 1 |
| chr3_-_95891938 | 0.18 |
ENSMUST00000036360.6
ENSMUST00000090476.3 |
BC028528
|
cDNA sequence BC028528 |
| chr12_+_85473883 | 0.18 |
ENSMUST00000021674.6
|
Fos
|
FBJ osteosarcoma oncogene |
| chr4_+_19280850 | 0.17 |
ENSMUST00000102999.1
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr2_-_60673654 | 0.17 |
ENSMUST00000059888.8
ENSMUST00000154764.1 |
Itgb6
|
integrin beta 6 |
| chr8_-_47675130 | 0.15 |
ENSMUST00000080353.2
|
Ing2
|
inhibitor of growth family, member 2 |
| chr4_-_122886044 | 0.15 |
ENSMUST00000106255.1
ENSMUST00000106257.3 |
Cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr17_-_46153517 | 0.15 |
ENSMUST00000171172.1
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr15_-_77842133 | 0.15 |
ENSMUST00000016771.6
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chrX_-_166518567 | 0.14 |
ENSMUST00000112187.1
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr7_+_17972124 | 0.14 |
ENSMUST00000094799.2
|
Ceacam11
|
carcinoembryonic antigen-related cell adhesion molecule 11 |
| chr15_+_11000716 | 0.13 |
ENSMUST00000117100.1
ENSMUST00000022851.7 |
Slc45a2
|
solute carrier family 45, member 2 |
| chrX_-_79434418 | 0.13 |
ENSMUST00000179788.1
|
Chdc2
|
calponin homology domain containing 2 |
| chr13_+_64432479 | 0.13 |
ENSMUST00000021939.6
|
Cdk20
|
cyclin-dependent kinase 20 |
| chr16_+_11313812 | 0.12 |
ENSMUST00000023140.5
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr14_-_6108665 | 0.12 |
ENSMUST00000165193.1
|
Gm3468
|
predicted gene 3468 |
| chr9_-_58313189 | 0.10 |
ENSMUST00000061799.8
|
Loxl1
|
lysyl oxidase-like 1 |
| chr14_+_20929416 | 0.10 |
ENSMUST00000022369.7
|
Vcl
|
vinculin |
| chr7_-_19310035 | 0.06 |
ENSMUST00000003640.2
|
Fosb
|
FBJ osteosarcoma oncogene B |
| chr7_+_19131686 | 0.05 |
ENSMUST00000165913.1
|
Fbxo46
|
F-box protein 46 |
| chr17_-_27204357 | 0.05 |
ENSMUST00000055117.7
|
Lemd2
|
LEM domain containing 2 |
| chr7_-_44997535 | 0.03 |
ENSMUST00000124232.1
ENSMUST00000003290.4 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr7_+_18009881 | 0.03 |
ENSMUST00000108488.1
ENSMUST00000081703.6 |
Ceacam13
|
carcinoembryonic antigen-related cell adhesion molecule 13 |
| chr2_-_17996924 | 0.03 |
ENSMUST00000105001.2
|
H2afb1
|
H2A histone family, member B1 |
| chr14_+_3049285 | 0.01 |
ENSMUST00000166494.1
|
Gm2897
|
predicted gene 2897 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 18.0 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.7 | 23.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.7 | 2.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.6 | 1.9 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.5 | 1.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.4 | 2.6 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.4 | 1.7 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 1.0 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.3 | 1.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.3 | 1.4 | GO:0071899 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.3 | 3.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.3 | 3.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 1.0 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.3 | 1.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.9 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.2 | 0.7 | GO:2000845 | positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 1.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.8 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 0.6 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.2 | 0.5 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.1 | GO:1903918 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.1 | 7.7 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 1.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.6 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.5 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 1.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 3.2 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 2.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.9 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 3.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 3.7 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 0.1 | 1.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.4 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.9 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.3 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.0 | 0.8 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 3.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.3 | 13.2 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 5.7 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 3.7 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 0.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 2.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 23.6 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.9 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.5 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.8 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 1.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.7 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 2.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 4.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 19.0 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 18.0 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.4 | 2.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.3 | 1.0 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.3 | 7.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.3 | 5.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.3 | 1.7 | GO:1902282 | phosphorelay sensor kinase activity(GO:0000155) voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.3 | 4.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 0.9 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.2 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 1.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.0 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 11.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 0.8 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 2.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.5 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 4.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 17.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 7.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 8.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 1.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.9 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 18.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 10.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 5.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.9 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |