Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Runx2_Bcl11a
Z-value: 3.82
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
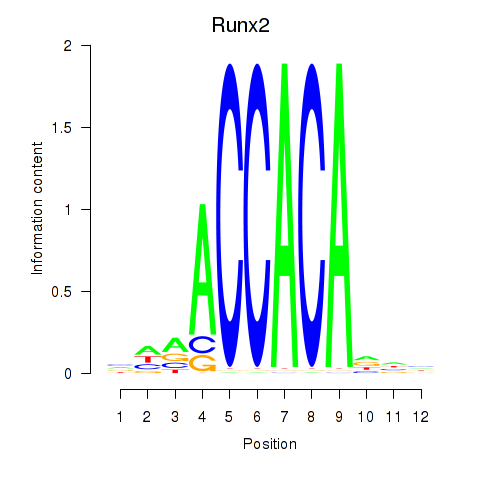
Runx2
|
ENSMUSG00000039153.10 | runt related transcription factor 2 |
|
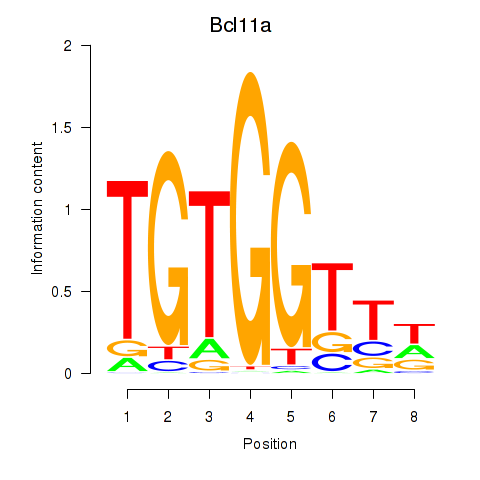
Bcl11a
|
ENSMUSG00000000861.9 | B cell CLL/lymphoma 11A (zinc finger protein) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl11a | mm10_v2_chr11_+_24078022_24078076 | 0.82 | 1.3e-09 | Click! |
| Runx2 | mm10_v2_chr17_-_44735612_44735710 | 0.78 | 2.0e-08 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_87875524 | 33.94 |
ENSMUST00000049768.3
|
Epx
|
eosinophil peroxidase |
| chr11_+_87793470 | 26.91 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr11_+_87793722 | 25.34 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr7_+_143005046 | 25.24 |
ENSMUST00000009396.6
|
Tspan32
|
tetraspanin 32 |
| chr4_-_46404224 | 21.61 |
ENSMUST00000107764.2
|
Hemgn
|
hemogen |
| chr18_-_35649349 | 20.89 |
ENSMUST00000025211.4
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr5_+_90768511 | 20.69 |
ENSMUST00000031319.6
|
Ppbp
|
pro-platelet basic protein |
| chr7_+_143005677 | 19.86 |
ENSMUST00000082008.5
ENSMUST00000105925.1 ENSMUST00000105924.1 |
Tspan32
|
tetraspanin 32 |
| chr7_+_143005638 | 19.77 |
ENSMUST00000075172.5
ENSMUST00000105923.1 |
Tspan32
|
tetraspanin 32 |
| chr7_-_142661858 | 18.83 |
ENSMUST00000145896.2
|
Igf2
|
insulin-like growth factor 2 |
| chr10_+_75564086 | 17.61 |
ENSMUST00000141062.1
ENSMUST00000152657.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr16_-_92826004 | 17.16 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr7_-_141016892 | 16.92 |
ENSMUST00000081924.3
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr14_-_56262233 | 16.72 |
ENSMUST00000015581.4
|
Gzmb
|
granzyme B |
| chr16_+_36277145 | 15.95 |
ENSMUST00000042097.9
|
Stfa1
|
stefin A1 |
| chr2_-_28084877 | 14.82 |
ENSMUST00000028179.8
ENSMUST00000117486.1 ENSMUST00000135472.1 |
Fcnb
|
ficolin B |
| chr7_-_142666816 | 14.25 |
ENSMUST00000105935.1
|
Igf2
|
insulin-like growth factor 2 |
| chr11_-_69617879 | 13.79 |
ENSMUST00000005334.2
|
Shbg
|
sex hormone binding globulin |
| chr17_+_33638056 | 13.13 |
ENSMUST00000052079.7
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr17_-_35085609 | 12.89 |
ENSMUST00000038507.6
|
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr7_-_103827922 | 12.59 |
ENSMUST00000023934.6
ENSMUST00000153218.1 |
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr8_-_11008458 | 11.92 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr4_+_115057410 | 11.82 |
ENSMUST00000136946.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr4_+_136172367 | 11.78 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr10_-_79788924 | 11.29 |
ENSMUST00000020573.6
|
Prss57
|
protease, serine 57 |
| chr13_-_19824234 | 11.22 |
ENSMUST00000065335.2
|
Gpr141
|
G protein-coupled receptor 141 |
| chr11_-_11970540 | 11.18 |
ENSMUST00000109653.1
|
Grb10
|
growth factor receptor bound protein 10 |
| chr3_-_59101810 | 10.77 |
ENSMUST00000085040.4
|
Gpr171
|
G protein-coupled receptor 171 |
| chr7_-_142576492 | 10.56 |
ENSMUST00000140716.1
|
H19
|
H19 fetal liver mRNA |
| chr16_+_17980565 | 10.19 |
ENSMUST00000075371.3
|
Vpreb2
|
pre-B lymphocyte gene 2 |
| chr9_+_8544196 | 10.12 |
ENSMUST00000050433.6
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr3_+_103860265 | 9.95 |
ENSMUST00000029433.7
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr5_-_107726017 | 9.94 |
ENSMUST00000159263.2
|
Gfi1
|
growth factor independent 1 |
| chrX_+_93654863 | 9.91 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chrX_+_159697308 | 9.62 |
ENSMUST00000123433.1
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr16_-_36367623 | 9.54 |
ENSMUST00000096089.2
|
BC100530
|
cDNA sequence BC100530 |
| chr1_+_135133272 | 9.26 |
ENSMUST00000167080.1
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr10_+_43579161 | 9.23 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr13_-_37050237 | 9.19 |
ENSMUST00000164727.1
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chrX_-_8090442 | 9.13 |
ENSMUST00000033505.6
|
Was
|
Wiskott-Aldrich syndrome homolog (human) |
| chr16_+_36156801 | 9.10 |
ENSMUST00000079184.4
|
Stfa2l1
|
stefin A2 like 1 |
| chr8_-_111691002 | 8.92 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr10_-_62379852 | 8.89 |
ENSMUST00000143236.1
ENSMUST00000133429.1 ENSMUST00000132926.1 ENSMUST00000116238.2 |
Hk1
|
hexokinase 1 |
| chr13_+_76579670 | 8.86 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_123123423 | 8.82 |
ENSMUST00000032248.7
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr6_+_123123313 | 8.73 |
ENSMUST00000041779.6
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_-_43533171 | 8.64 |
ENSMUST00000004728.5
ENSMUST00000039861.5 |
Cd33
|
CD33 antigen |
| chr7_+_44572370 | 8.52 |
ENSMUST00000002274.8
|
Napsa
|
napsin A aspartic peptidase |
| chr7_-_25005895 | 8.50 |
ENSMUST00000102858.3
ENSMUST00000080882.6 |
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr19_-_20390944 | 8.50 |
ENSMUST00000025561.7
|
Anxa1
|
annexin A1 |
| chr7_-_100863373 | 8.35 |
ENSMUST00000142885.1
ENSMUST00000008462.3 |
Relt
|
RELT tumor necrosis factor receptor |
| chr4_-_137430517 | 8.09 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr1_+_40515362 | 8.03 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr7_-_14254870 | 8.01 |
ENSMUST00000184731.1
ENSMUST00000076576.6 |
Sult2a6
|
sulfotransferase family 2A, dehydroepiandrosterone (DHEA)-preferring, member 6 |
| chr2_-_26021679 | 7.94 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr11_-_83649349 | 7.90 |
ENSMUST00000001008.5
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr2_-_32387760 | 7.86 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr4_+_135120640 | 7.85 |
ENSMUST00000056977.7
|
Runx3
|
runt related transcription factor 3 |
| chr2_-_26021532 | 7.75 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr17_+_35821675 | 7.70 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr5_+_90772435 | 7.56 |
ENSMUST00000031320.6
|
Pf4
|
platelet factor 4 |
| chrX_-_136068236 | 7.51 |
ENSMUST00000049130.7
|
Bex2
|
brain expressed X-linked 2 |
| chr15_+_44457522 | 7.49 |
ENSMUST00000166957.1
ENSMUST00000038336.5 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr12_-_4874341 | 7.32 |
ENSMUST00000137337.1
ENSMUST00000045921.7 |
Mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr7_+_24370255 | 7.10 |
ENSMUST00000171904.1
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr14_+_80000292 | 7.02 |
ENSMUST00000088735.3
|
Olfm4
|
olfactomedin 4 |
| chr4_-_43499608 | 6.94 |
ENSMUST00000136005.1
ENSMUST00000054538.6 |
Arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr11_-_79523760 | 6.87 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr8_+_88294204 | 6.87 |
ENSMUST00000098521.2
|
Adcy7
|
adenylate cyclase 7 |
| chr2_-_58160495 | 6.79 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr17_+_36869567 | 6.76 |
ENSMUST00000060524.9
|
Trim10
|
tripartite motif-containing 10 |
| chr7_-_142656018 | 6.66 |
ENSMUST00000178921.1
|
Igf2
|
insulin-like growth factor 2 |
| chr17_-_35066170 | 6.55 |
ENSMUST00000174190.1
ENSMUST00000097337.1 |
AU023871
|
expressed sequence AU023871 |
| chr13_+_112288451 | 6.44 |
ENSMUST00000022275.6
ENSMUST00000056047.7 ENSMUST00000165593.1 |
Ankrd55
|
ankyrin repeat domain 55 |
| chr4_+_115057683 | 6.44 |
ENSMUST00000161601.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr10_+_75571522 | 6.38 |
ENSMUST00000143226.1
ENSMUST00000124259.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr6_-_129917650 | 6.30 |
ENSMUST00000118060.1
|
Klra5
|
killer cell lectin-like receptor, subfamily A, member 5 |
| chr11_-_72550255 | 6.27 |
ENSMUST00000021154.6
|
Spns3
|
spinster homolog 3 |
| chr17_+_47596061 | 6.19 |
ENSMUST00000182539.1
|
Ccnd3
|
cyclin D3 |
| chr2_-_172370506 | 6.17 |
ENSMUST00000109139.1
ENSMUST00000028997.7 ENSMUST00000109140.3 |
Aurka
|
aurora kinase A |
| chr3_-_15332285 | 6.09 |
ENSMUST00000108361.1
|
Gm9733
|
predicted gene 9733 |
| chr6_-_40585783 | 6.08 |
ENSMUST00000177178.1
ENSMUST00000129948.2 ENSMUST00000101491.4 |
Clec5a
|
C-type lectin domain family 5, member a |
| chr6_-_87533219 | 6.04 |
ENSMUST00000113637.2
ENSMUST00000071024.6 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr2_-_28699661 | 5.98 |
ENSMUST00000124840.1
|
1700026L06Rik
|
RIKEN cDNA 1700026L06 gene |
| chr15_-_79285502 | 5.97 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr14_-_60086832 | 5.88 |
ENSMUST00000080368.5
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr11_+_3330781 | 5.87 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr15_-_66801577 | 5.85 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr7_+_143005770 | 5.83 |
ENSMUST00000143512.1
|
Tspan32
|
tetraspanin 32 |
| chr15_-_103252810 | 5.81 |
ENSMUST00000154510.1
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr2_-_28699636 | 5.74 |
ENSMUST00000102877.1
|
1700026L06Rik
|
RIKEN cDNA 1700026L06 gene |
| chr19_-_4615453 | 5.68 |
ENSMUST00000053597.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr3_+_146150174 | 5.63 |
ENSMUST00000098524.4
|
Mcoln2
|
mucolipin 2 |
| chr14_-_56085214 | 5.62 |
ENSMUST00000015594.7
|
Mcpt8
|
mast cell protease 8 |
| chr1_+_135132693 | 5.57 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr17_-_13404191 | 5.53 |
ENSMUST00000115650.1
|
Gm8597
|
predicted gene 8597 |
| chr14_+_55853997 | 5.48 |
ENSMUST00000100529.3
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chrX_-_57338598 | 5.48 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr3_+_96181151 | 5.48 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr14_+_55765956 | 5.46 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr15_-_103255433 | 5.46 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr15_-_9529868 | 5.36 |
ENSMUST00000003981.4
|
Il7r
|
interleukin 7 receptor |
| chr15_-_78493607 | 5.33 |
ENSMUST00000163494.1
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr11_-_106314494 | 5.24 |
ENSMUST00000167143.1
|
Cd79b
|
CD79B antigen |
| chr1_-_38664947 | 5.18 |
ENSMUST00000039827.7
ENSMUST00000027250.7 |
Aff3
|
AF4/FMR2 family, member 3 |
| chr2_+_172393900 | 5.17 |
ENSMUST00000109136.2
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr15_-_79285470 | 5.00 |
ENSMUST00000170955.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr1_+_170277376 | 5.00 |
ENSMUST00000179976.1
|
Sh2d1b1
|
SH2 domain protein 1B1 |
| chr17_-_28560704 | 4.98 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr9_+_53405280 | 4.98 |
ENSMUST00000005262.1
|
4930550C14Rik
|
RIKEN cDNA 4930550C14 gene |
| chr6_+_86628174 | 4.95 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr14_+_66297029 | 4.92 |
ENSMUST00000022623.6
ENSMUST00000121006.1 |
Trim35
|
tripartite motif-containing 35 |
| chr14_+_27000362 | 4.78 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr5_+_122100951 | 4.75 |
ENSMUST00000014080.6
ENSMUST00000111750.1 ENSMUST00000139213.1 ENSMUST00000111751.1 ENSMUST00000155612.1 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr4_-_92191749 | 4.71 |
ENSMUST00000123179.1
|
Gm12666
|
predicted gene 12666 |
| chr11_+_31832660 | 4.70 |
ENSMUST00000132857.1
|
Gm12107
|
predicted gene 12107 |
| chr14_+_56042123 | 4.67 |
ENSMUST00000015576.4
|
Mcpt2
|
mast cell protease 2 |
| chr17_-_35027909 | 4.66 |
ENSMUST00000040151.2
|
Sapcd1
|
suppressor APC domain containing 1 |
| chr4_+_127172866 | 4.66 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr16_-_16869255 | 4.66 |
ENSMUST00000075017.4
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr6_+_87778084 | 4.64 |
ENSMUST00000032133.3
|
Gp9
|
glycoprotein 9 (platelet) |
| chr7_+_30776394 | 4.62 |
ENSMUST00000041703.7
|
Dmkn
|
dermokine |
| chr13_-_22219820 | 4.55 |
ENSMUST00000057516.1
|
Vmn1r193
|
vomeronasal 1 receptor 193 |
| chr1_-_144775419 | 4.50 |
ENSMUST00000027603.3
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr19_-_4615647 | 4.50 |
ENSMUST00000113822.2
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4 |
| chr12_-_8539545 | 4.45 |
ENSMUST00000095863.3
ENSMUST00000165657.1 |
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr12_-_112860886 | 4.41 |
ENSMUST00000021729.7
|
Gpr132
|
G protein-coupled receptor 132 |
| chr10_-_62507737 | 4.38 |
ENSMUST00000020271.6
|
Srgn
|
serglycin |
| chr4_-_134012381 | 4.36 |
ENSMUST00000176113.1
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_+_43437073 | 4.32 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr19_-_5273080 | 4.29 |
ENSMUST00000025786.7
|
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr18_-_62179948 | 4.26 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr11_-_102088471 | 4.19 |
ENSMUST00000017458.4
|
Mpp2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) |
| chr10_-_117792663 | 4.14 |
ENSMUST00000167943.1
ENSMUST00000064848.5 |
Nup107
|
nucleoporin 107 |
| chr15_-_75567176 | 4.12 |
ENSMUST00000156032.1
ENSMUST00000127095.1 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr5_-_122002340 | 4.11 |
ENSMUST00000134326.1
|
Cux2
|
cut-like homeobox 2 |
| chr2_-_73485733 | 4.07 |
ENSMUST00000102680.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr5_+_105519388 | 4.07 |
ENSMUST00000067924.6
ENSMUST00000150981.1 |
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr7_-_141655319 | 4.03 |
ENSMUST00000062451.7
|
Muc6
|
mucin 6, gastric |
| chr13_-_63398167 | 4.01 |
ENSMUST00000160735.1
|
Fancc
|
Fanconi anemia, complementation group C |
| chr2_-_73486456 | 3.99 |
ENSMUST00000141264.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_-_164935522 | 3.96 |
ENSMUST00000027860.7
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr17_+_12119274 | 3.92 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr15_+_9436028 | 3.92 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr9_-_95845215 | 3.91 |
ENSMUST00000093800.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr11_+_3330401 | 3.90 |
ENSMUST00000045153.4
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr5_+_66676098 | 3.88 |
ENSMUST00000031131.9
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1 |
| chr6_-_40951826 | 3.87 |
ENSMUST00000073642.5
|
Gm4744
|
predicted gene 4744 |
| chr1_-_89933290 | 3.87 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr17_+_8849974 | 3.86 |
ENSMUST00000115720.1
|
Pde10a
|
phosphodiesterase 10A |
| chr7_+_101361997 | 3.86 |
ENSMUST00000133423.1
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr3_-_54915867 | 3.84 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr11_-_17008647 | 3.83 |
ENSMUST00000102881.3
|
Plek
|
pleckstrin |
| chr9_+_51213683 | 3.82 |
ENSMUST00000034554.7
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr2_-_28563362 | 3.78 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr10_+_118441044 | 3.77 |
ENSMUST00000068592.3
|
Ifng
|
interferon gamma |
| chr5_+_37047464 | 3.77 |
ENSMUST00000137019.1
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr9_+_111004811 | 3.74 |
ENSMUST00000080872.4
|
Gm10030
|
predicted gene 10030 |
| chr11_-_109472611 | 3.64 |
ENSMUST00000168740.1
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr7_+_28540863 | 3.63 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr10_-_62508097 | 3.59 |
ENSMUST00000159020.1
|
Srgn
|
serglycin |
| chr8_-_107065632 | 3.57 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr13_+_76579681 | 3.54 |
ENSMUST00000109589.2
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr7_+_43351378 | 3.48 |
ENSMUST00000012798.7
ENSMUST00000122423.1 ENSMUST00000121494.1 |
Siglec5
|
sialic acid binding Ig-like lectin 5 |
| chr5_-_117319242 | 3.48 |
ENSMUST00000100834.1
|
Gm10399
|
predicted gene 10399 |
| chr18_+_62180119 | 3.47 |
ENSMUST00000067743.1
|
Gm9949
|
predicted gene 9949 |
| chr8_+_60655540 | 3.44 |
ENSMUST00000034066.3
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr19_-_4305955 | 3.44 |
ENSMUST00000025791.5
|
Adrbk1
|
adrenergic receptor kinase, beta 1 |
| chr8_-_68121527 | 3.44 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr12_+_116077720 | 3.43 |
ENSMUST00000011315.3
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr6_+_49036518 | 3.41 |
ENSMUST00000031840.7
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr2_-_170427828 | 3.40 |
ENSMUST00000013667.2
ENSMUST00000109152.2 ENSMUST00000068137.4 |
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr16_+_32756336 | 3.40 |
ENSMUST00000135753.1
|
Muc4
|
mucin 4 |
| chr5_+_139406387 | 3.34 |
ENSMUST00000052176.8
|
C130050O18Rik
|
RIKEN cDNA C130050O18 gene |
| chr5_+_37050854 | 3.33 |
ENSMUST00000043794.4
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr17_-_48146306 | 3.31 |
ENSMUST00000063481.7
|
9830107B12Rik
|
RIKEN cDNA 9830107B12 gene |
| chr17_+_29093763 | 3.29 |
ENSMUST00000023829.6
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr15_-_36555556 | 3.24 |
ENSMUST00000161202.1
ENSMUST00000013755.5 |
Snx31
|
sorting nexin 31 |
| chr5_+_86071734 | 3.23 |
ENSMUST00000031171.7
|
Stap1
|
signal transducing adaptor family member 1 |
| chr11_-_100207507 | 3.21 |
ENSMUST00000007272.7
|
Krt14
|
keratin 14 |
| chr7_-_3825641 | 3.20 |
ENSMUST00000094911.4
ENSMUST00000108619.1 ENSMUST00000108620.1 |
Gm15448
|
predicted gene 15448 |
| chr7_+_127211608 | 3.19 |
ENSMUST00000032910.6
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr13_+_113317084 | 3.16 |
ENSMUST00000136755.2
|
BC067074
|
cDNA sequence BC067074 |
| chr16_-_58718724 | 3.15 |
ENSMUST00000089318.3
|
Gpr15
|
G protein-coupled receptor 15 |
| chr2_+_25423234 | 3.13 |
ENSMUST00000134259.1
ENSMUST00000100320.4 |
Fut7
|
fucosyltransferase 7 |
| chr15_+_39076885 | 3.12 |
ENSMUST00000067072.3
|
Cthrc1
|
collagen triple helix repeat containing 1 |
| chr16_-_10395438 | 3.09 |
ENSMUST00000115831.1
|
Tekt5
|
tektin 5 |
| chr1_+_165302625 | 3.08 |
ENSMUST00000111450.1
|
Gpr161
|
G protein-coupled receptor 161 |
| chr14_-_51146757 | 3.07 |
ENSMUST00000080126.2
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr14_-_54686060 | 3.05 |
ENSMUST00000125265.1
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr13_-_113100971 | 3.04 |
ENSMUST00000023897.5
|
Gzma
|
granzyme A |
| chr7_-_45239041 | 3.01 |
ENSMUST00000131290.1
|
Cd37
|
CD37 antigen |
| chr7_+_121865070 | 3.00 |
ENSMUST00000033161.5
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chrX_+_73892102 | 3.00 |
ENSMUST00000033765.7
ENSMUST00000101470.2 ENSMUST00000114395.1 |
Avpr2
|
arginine vasopressin receptor 2 |
| chr11_-_76577701 | 2.99 |
ENSMUST00000176179.1
|
Abr
|
active BCR-related gene |
| chr11_-_100414829 | 2.99 |
ENSMUST00000066489.6
|
Leprel4
|
leprecan-like 4 |
| chrX_+_106027300 | 2.98 |
ENSMUST00000055941.6
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr14_-_19977151 | 2.97 |
ENSMUST00000055100.7
ENSMUST00000162425.1 |
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr9_-_95815389 | 2.97 |
ENSMUST00000119760.1
|
Pls1
|
plastin 1 (I-isoform) |
| chr6_-_128826305 | 2.94 |
ENSMUST00000174544.1
ENSMUST00000172887.1 ENSMUST00000032472.4 |
Klrb1b
|
killer cell lectin-like receptor subfamily B member 1B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.4 | 52.3 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 11.4 | 34.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 7.9 | 79.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 6.1 | 18.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 5.2 | 15.5 | GO:0071663 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 5.0 | 39.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 4.8 | 14.4 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 4.2 | 20.9 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 3.7 | 14.8 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 3.3 | 16.7 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 3.1 | 9.2 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 2.9 | 23.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 2.8 | 8.5 | GO:0036292 | DNA rewinding(GO:0036292) |
| 2.0 | 8.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 2.0 | 8.0 | GO:0033382 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 2.0 | 9.9 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 1.8 | 8.8 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 1.7 | 11.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 1.6 | 16.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 1.6 | 3.2 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 1.6 | 1.6 | GO:0045401 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 1.5 | 6.2 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 1.5 | 9.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 1.5 | 7.6 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 1.4 | 10.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 1.4 | 4.2 | GO:1904959 | elastin biosynthetic process(GO:0051542) regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.4 | 4.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.3 | 5.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 1.3 | 10.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 1.3 | 3.8 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 1.3 | 5.0 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 1.2 | 22.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 1.2 | 4.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 1.1 | 7.9 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 1.1 | 8.9 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 1.1 | 7.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 1.1 | 12.9 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 1.0 | 6.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.0 | 2.1 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 1.0 | 11.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 1.0 | 3.9 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.0 | 2.9 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.9 | 11.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.9 | 2.7 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.9 | 2.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 2.7 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.9 | 7.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.9 | 2.7 | GO:0009726 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.9 | 2.7 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.8 | 2.5 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.8 | 10.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.8 | 4.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.8 | 2.5 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.8 | 5.9 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.8 | 3.4 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.8 | 2.5 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.8 | 8.3 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.8 | 2.5 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.8 | 2.4 | GO:0070843 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.8 | 10.6 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.8 | 3.2 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.8 | 3.8 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.7 | 3.7 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.7 | 2.2 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.7 | 2.9 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.7 | 2.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.7 | 2.1 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.7 | 9.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.7 | 4.2 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.7 | 2.7 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.7 | 8.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.7 | 5.3 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.7 | 11.9 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.7 | 2.0 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.6 | 3.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 1.9 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.6 | 2.5 | GO:2000612 | regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.6 | 1.8 | GO:0034635 | glutathione transport(GO:0034635) oligopeptide transmembrane transport(GO:0035672) tripeptide transport(GO:0042939) |
| 0.6 | 0.6 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.6 | 1.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.6 | 11.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.6 | 1.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.6 | 0.6 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.5 | 1.6 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.5 | 2.2 | GO:0046722 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.5 | 2.6 | GO:0046110 | xanthine catabolic process(GO:0009115) xanthine metabolic process(GO:0046110) |
| 0.5 | 2.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.5 | 2.0 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.5 | 4.4 | GO:1901724 | miRNA catabolic process(GO:0010587) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.5 | 1.9 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.5 | 3.9 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.5 | 4.8 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.5 | 1.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.5 | 1.4 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.5 | 0.9 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.5 | 1.9 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.5 | 3.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 1.4 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.4 | 1.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.4 | 10.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 3.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.4 | 4.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.4 | 8.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.4 | 2.5 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.4 | 2.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.4 | 5.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.4 | 1.2 | GO:0019046 | viral latency(GO:0019042) release from viral latency(GO:0019046) |
| 0.4 | 1.6 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.4 | 2.7 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.4 | 0.8 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.4 | 4.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.4 | 5.4 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.4 | 3.5 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.4 | 4.5 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.4 | 1.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 3.4 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.4 | 5.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.4 | 2.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.4 | 0.7 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.4 | 6.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 2.5 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.4 | 1.8 | GO:0043985 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) histone H4-R3 methylation(GO:0043985) |
| 0.4 | 2.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.3 | 2.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.3 | 2.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.3 | 2.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.3 | 5.0 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.3 | 0.7 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.3 | 7.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.3 | 5.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.3 | 1.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.3 | 1.3 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 1.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.3 | 0.6 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.3 | 5.0 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.3 | 0.9 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.3 | 2.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 1.8 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 | 0.3 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.3 | 1.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 3.0 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.3 | 6.1 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.3 | 4.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.3 | 0.9 | GO:0061642 | chemoattraction of axon(GO:0061642) inner medullary collecting duct development(GO:0072061) |
| 0.3 | 2.0 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.3 | 0.8 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.3 | 1.4 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.3 | 3.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 0.5 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.3 | 1.1 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 | 1.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 | 1.9 | GO:0042148 | strand invasion(GO:0042148) |
| 0.3 | 0.8 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.3 | 2.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.3 | 4.8 | GO:0002716 | negative regulation of natural killer cell mediated immunity(GO:0002716) |
| 0.3 | 3.0 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.2 | 1.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.9 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.2 | 1.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.2 | 3.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 | 0.5 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.2 | 1.4 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.2 | 1.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.2 | 0.5 | GO:0001802 | type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.2 | 1.4 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 7.9 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.2 | 7.0 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.2 | 0.7 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.2 | 4.9 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 6.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.2 | 2.3 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 2.7 | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.2 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.7 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.2 | 0.7 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 1.7 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 0.4 | GO:0042322 | negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) |
| 0.2 | 0.2 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.2 | 1.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 2.7 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.2 | 2.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 2.4 | GO:0021779 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.2 | 2.8 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 2.0 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.2 | 5.3 | GO:0032967 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.2 | 0.8 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 5.4 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.2 | 18.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.2 | 1.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.2 | 0.9 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 0.7 | GO:0046726 | regulation by virus of viral protein levels in host cell(GO:0046719) positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 | 0.3 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.2 | 3.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.2 | 3.0 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.2 | 1.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 0.8 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.2 | 0.5 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.2 | 0.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 0.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 5.0 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 1.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.4 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 1.3 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.1 | 0.7 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 1.3 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.4 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.1 | 0.6 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.6 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 2.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.6 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 3.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 5.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.1 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.1 | 0.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 1.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.9 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.7 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 2.7 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.5 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 | 1.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.1 | 0.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.0 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.1 | 4.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 1.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 2.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 2.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 2.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.5 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 1.9 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.6 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 | 4.7 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 2.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.3 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.6 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 | 14.9 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 4.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.5 | GO:0034627 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.7 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.1 | 1.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.7 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 1.1 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 2.9 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.5 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.1 | 0.5 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.8 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.1 | 5.4 | GO:0033619 | membrane protein proteolysis(GO:0033619) |
| 0.1 | 2.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.8 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.3 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 2.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.1 | 4.0 | GO:0035137 | hindlimb morphogenesis(GO:0035137) |
| 0.1 | 1.0 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.8 | GO:0070571 | negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 2.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0021664 | rhombomere morphogenesis(GO:0021593) rhombomere formation(GO:0021594) rhombomere 3 morphogenesis(GO:0021658) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 1.2 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 1.7 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 4.7 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.1 | GO:0050718 | regulation of interleukin-1 secretion(GO:0050704) regulation of interleukin-1 beta secretion(GO:0050706) positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 0.7 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.1 | 0.4 | GO:0072366 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.5 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:0015822 | ornithine transport(GO:0015822) |
| 0.1 | 4.8 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 0.5 | GO:0071816 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 0.1 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) monocyte aggregation(GO:0070487) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 1.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.7 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 3.3 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.2 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 | 1.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.4 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 7.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.7 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.0 | GO:0032674 | regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 2.1 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.1 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.1 | 0.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.0 | 0.7 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 1.7 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.9 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 6.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.9 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.5 | GO:0044036 | cell wall macromolecule catabolic process(GO:0016998) cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.9 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.3 | GO:0050672 | negative regulation of mononuclear cell proliferation(GO:0032945) negative regulation of T cell proliferation(GO:0042130) negative regulation of lymphocyte proliferation(GO:0050672) |
| 0.0 | 1.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 1.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0002418 | immune response to tumor cell(GO:0002418) |
| 0.0 | 0.6 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.8 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.7 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 1.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 1.7 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.8 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 1.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.9 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 1.2 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 1.3 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.0 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) |
| 0.0 | 0.2 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 2.7 | GO:0022412 | cellular process involved in reproduction in multicellular organism(GO:0022412) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.7 | 70.7 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 6.1 | 18.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 4.2 | 16.8 | GO:0044194 | cytolytic granule(GO:0044194) |
| 4.0 | 63.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 2.1 | 6.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 1.8 | 5.3 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 1.7 | 8.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.5 | 16.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 1.1 | 3.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.1 | 15.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 1.0 | 11.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.9 | 2.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.9 | 8.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.9 | 10.4 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.9 | 6.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.8 | 2.5 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.7 | 3.0 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.7 | 2.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 7.0 | GO:0042581 | specific granule(GO:0042581) |
| 0.7 | 4.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.7 | 17.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.7 | 7.4 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.6 | 2.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.6 | 1.8 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.6 | 1.7 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.6 | 0.6 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.6 | 2.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.5 | 2.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.5 | 2.0 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.5 | 6.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.5 | 1.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 7.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.4 | 4.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.4 | 4.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 3.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 2.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.4 | 1.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.4 | 4.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 0.9 | GO:1990047 | spindle matrix(GO:1990047) |
| 0.3 | 9.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 3.9 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.3 | 1.2 | GO:0090537 | CERF complex(GO:0090537) |
| 0.3 | 0.8 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 2.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 3.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 0.8 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.2 | 0.7 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.2 | 1.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 2.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.7 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.2 | 4.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 2.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.2 | 0.8 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.2 | 9.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 2.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.9 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.2 | 1.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 3.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 10.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 4.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 2.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 0.9 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 0.6 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.2 | 1.4 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.2 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 9.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 3.0 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 7.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 6.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 0.9 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 26.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 2.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.1 | 1.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 2.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 12.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 0.8 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 2.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 6.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 6.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.1 | 2.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 14.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 10.6 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.8 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.3 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.7 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 2.9 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.5 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.1 | 12.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 1.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 11.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.1 | 1.7 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.2 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.1 | 1.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.7 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.1 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 7.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.7 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 13.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 2.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 6.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 4.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 13.2 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.6 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 6.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 56.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 1.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 7.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 2.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 4.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.8 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.0 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 12.6 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 2.4 | 9.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 2.2 | 28.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 2.0 | 8.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 2.0 | 23.9 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 2.0 | 9.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 1.8 | 16.0 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.8 | 7.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.7 | 8.5 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 1.6 | 14.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.6 | 4.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 1.5 | 1.5 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 1.4 | 4.2 | GO:0045353 | interleukin-1, Type II receptor binding(GO:0005151) interleukin-1 Type I receptor antagonist activity(GO:0045352) interleukin-1 Type II receptor antagonist activity(GO:0045353) |
| 1.4 | 79.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 1.4 | 5.5 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 1.3 | 3.9 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.3 | 3.8 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 1.3 | 3.8 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 1.2 | 9.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 1.1 | 8.0 | GO:0004027 | alcohol sulfotransferase activity(GO:0004027) |
| 1.1 | 3.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.1 | 3.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 1.1 | 4.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.1 | 62.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 1.1 | 4.2 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.9 | 2.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.9 | 2.7 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.9 | 1.7 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.9 | 2.6 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.8 | 3.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.8 | 10.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.8 | 2.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.8 | 2.3 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.8 | 8.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.7 | 5.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.7 | 8.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.7 | 2.1 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.7 | 10.1 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.6 | 10.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.6 | 3.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 2.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.6 | 2.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.6 | 9.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 2.6 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.5 | 3.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.5 | 1.6 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.5 | 3.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.5 | 3.0 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.5 | 13.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.5 | 1.4 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.5 | 1.4 | GO:0051379 | alpha2-adrenergic receptor activity(GO:0004938) epinephrine binding(GO:0051379) |
| 0.5 | 4.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 5.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.5 | 1.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.4 | 3.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 3.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.4 | 3.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 2.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 2.9 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 11.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.4 | 2.0 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.4 | 2.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 1.6 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.4 | 1.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 1.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.4 | 1.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.4 | 4.9 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.4 | 2.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 2.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 7.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 15.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 2.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 2.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 19.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.3 | 3.0 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.3 | 5.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.3 | 0.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 2.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.3 | 1.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 2.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.3 | 8.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.3 | 1.8 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.3 | 9.7 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 4.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.3 | 5.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 4.5 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 2.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.3 | 1.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 6.1 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.3 | 1.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 0.8 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.3 | 1.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 5.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 6.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.2 | GO:0052794 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 0.7 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.2 | 4.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.9 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 2.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 7.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 2.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 0.8 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 0.6 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.2 | 2.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 9.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 0.9 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.2 | 3.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 2.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 0.9 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 1.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.2 | 4.8 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.2 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 2.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.2 | 1.5 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 0.7 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.7 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.2 | 1.7 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 4.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 21.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 0.9 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 1.5 | GO:0052744 | phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.2 | 7.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.2 | 4.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 0.9 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 2.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.9 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 2.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.5 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.1 | 2.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 24.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 3.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 7.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 14.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.5 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.7 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.1 | 0.5 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 2.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 17.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 6.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.0 | GO:0016505 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.1 | 4.9 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 4.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 1.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.8 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.3 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) TIR domain binding(GO:0070976) |
| 0.1 | 1.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 2.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.4 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0035586 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) purinergic receptor activity(GO:0035586) |
| 0.1 | 5.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 10.2 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 4.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 2.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 5.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.9 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 0.1 | 0.1 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 3.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 8.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 5.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 2.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 1.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0030169 | low-density lipoprotein receptor activity(GO:0005041) low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 5.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.1 | 15.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.8 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 11.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.3 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 6.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 2.6 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.7 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0034711 | inhibin binding(GO:0034711) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 2.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 4.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 1.7 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 3.4 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 3.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) U6 snRNA 3'-end binding(GO:0030629) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 3.0 | GO:0061135 | endopeptidase inhibitor activity(GO:0004866) endopeptidase regulator activity(GO:0061135) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 5.1 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 3.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 60.8 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.8 | 14.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.8 | 13.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.7 | 23.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.6 | 23.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.6 | 34.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.5 | 36.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.5 | 9.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.4 | 22.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.4 | 14.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.4 | 9.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.4 | 7.5 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.4 | 9.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.3 | 1.9 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 7.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 2.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 7.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.2 | 1.9 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 9.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 5.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 6.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 5.6 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 2.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 1.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 4.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.2 | 3.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 5.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 5.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 13.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 4.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 12.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 2.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 7.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 19.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 6.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.0 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.1 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 0.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.0 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 2.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 6.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 2.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 36.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.2 | 16.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 1.1 | 10.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 1.1 | 22.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 1.0 | 11.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.9 | 2.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.7 | 33.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.7 | 9.8 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.4 | 15.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.4 | 6.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.4 | 17.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.4 | 4.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.4 | 16.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 4.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 9.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.3 | 6.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 0.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 13.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.3 | 5.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.3 | 3.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 3.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.2 | 5.7 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 10.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.2 | 7.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.2 | 10.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 2.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.2 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 9.1 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.2 | 1.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 4.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 4.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 8.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 4.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.2 | 3.9 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.2 | 9.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 4.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.7 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 3.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 3.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 2.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 4.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 9.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 3.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 3.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 25.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.0 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 2.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 5.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 3.9 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 7.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 15.3 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.1 | 5.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 4.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 3.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.1 | 2.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 2.1 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.1 | 3.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 4.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 3.0 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 0.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.8 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 4.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 3.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 2.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.8 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 2.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 2.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |