Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
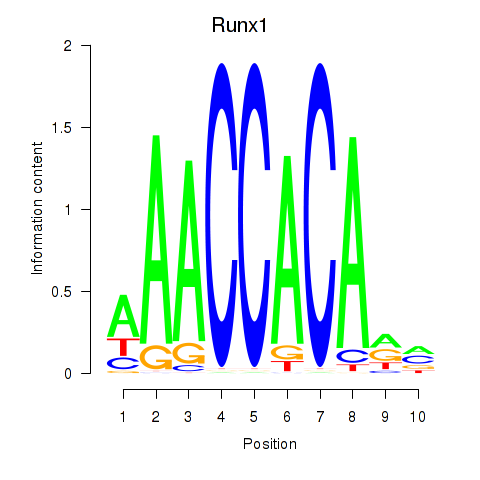
Results for Runx1
Z-value: 1.98
Transcription factors associated with Runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx1
|
ENSMUSG00000022952.10 | runt related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx1 | mm10_v2_chr16_-_92826004_92826080 | 0.87 | 5.0e-12 | Click! |
Activity profile of Runx1 motif
Sorted Z-values of Runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87793470 | 15.27 |
ENSMUST00000020779.4
|
Mpo
|
myeloperoxidase |
| chr11_+_87793722 | 14.45 |
ENSMUST00000143021.2
|
Mpo
|
myeloperoxidase |
| chr16_-_92826004 | 13.22 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr2_-_28084877 | 10.65 |
ENSMUST00000028179.8
ENSMUST00000117486.1 ENSMUST00000135472.1 |
Fcnb
|
ficolin B |
| chr4_-_46404224 | 10.13 |
ENSMUST00000107764.2
|
Hemgn
|
hemogen |
| chr15_-_103255433 | 8.50 |
ENSMUST00000075192.6
|
Nfe2
|
nuclear factor, erythroid derived 2 |
| chr3_-_59101810 | 7.26 |
ENSMUST00000085040.4
|
Gpr171
|
G protein-coupled receptor 171 |
| chr1_+_40515362 | 6.96 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr5_-_107726017 | 6.87 |
ENSMUST00000159263.2
|
Gfi1
|
growth factor independent 1 |
| chr2_+_164948219 | 6.76 |
ENSMUST00000017881.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr13_-_113100971 | 6.43 |
ENSMUST00000023897.5
|
Gzma
|
granzyme A |
| chr7_+_143005046 | 6.02 |
ENSMUST00000009396.6
|
Tspan32
|
tetraspanin 32 |
| chr7_-_127137807 | 5.84 |
ENSMUST00000049931.5
|
Spn
|
sialophorin |
| chr14_+_56042123 | 5.78 |
ENSMUST00000015576.4
|
Mcpt2
|
mast cell protease 2 |
| chr7_+_44572370 | 5.63 |
ENSMUST00000002274.8
|
Napsa
|
napsin A aspartic peptidase |
| chr7_+_24370255 | 5.49 |
ENSMUST00000171904.1
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr13_+_76579670 | 5.31 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr15_-_66801577 | 5.25 |
ENSMUST00000168589.1
|
Sla
|
src-like adaptor |
| chr2_+_172393900 | 5.10 |
ENSMUST00000109136.2
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr17_+_31208049 | 5.00 |
ENSMUST00000173776.1
|
Ubash3a
|
ubiquitin associated and SH3 domain containing, A |
| chrX_-_8090442 | 4.34 |
ENSMUST00000033505.6
|
Was
|
Wiskott-Aldrich syndrome homolog (human) |
| chr18_-_35649349 | 4.32 |
ENSMUST00000025211.4
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr2_-_58160495 | 4.29 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr4_+_135120640 | 3.89 |
ENSMUST00000056977.7
|
Runx3
|
runt related transcription factor 3 |
| chr3_+_103860265 | 3.89 |
ENSMUST00000029433.7
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
| chr8_+_105690906 | 3.79 |
ENSMUST00000062574.6
|
Rltpr
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing |
| chr17_+_47596061 | 3.62 |
ENSMUST00000182539.1
|
Ccnd3
|
cyclin D3 |
| chr17_+_12119274 | 3.25 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr7_-_43533171 | 3.19 |
ENSMUST00000004728.5
ENSMUST00000039861.5 |
Cd33
|
CD33 antigen |
| chr2_-_26021679 | 3.07 |
ENSMUST00000036509.7
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr11_-_83649349 | 3.03 |
ENSMUST00000001008.5
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chrX_-_57338598 | 3.01 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_133872997 | 2.97 |
ENSMUST00000137486.2
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr2_-_26021532 | 2.96 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr12_+_85686648 | 2.95 |
ENSMUST00000040536.5
|
Batf
|
basic leucine zipper transcription factor, ATF-like |
| chr14_-_56262233 | 2.87 |
ENSMUST00000015581.4
|
Gzmb
|
granzyme B |
| chr7_-_25132473 | 2.84 |
ENSMUST00000108418.4
ENSMUST00000108415.3 ENSMUST00000098679.3 ENSMUST00000175774.2 ENSMUST00000108417.3 ENSMUST00000108416.3 ENSMUST00000108414.1 ENSMUST00000108413.1 ENSMUST00000176408.1 |
Pou2f2
|
POU domain, class 2, transcription factor 2 |
| chr11_+_69966896 | 2.69 |
ENSMUST00000151515.1
|
Cldn7
|
claudin 7 |
| chr8_-_11008458 | 2.67 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr3_+_96181151 | 2.54 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr17_+_33638056 | 2.48 |
ENSMUST00000052079.7
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1 |
| chr13_+_76579681 | 2.41 |
ENSMUST00000109589.2
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr6_-_136941494 | 2.38 |
ENSMUST00000111892.1
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr15_-_9529868 | 2.38 |
ENSMUST00000003981.4
|
Il7r
|
interleukin 7 receptor |
| chr11_-_79530569 | 2.35 |
ENSMUST00000103236.3
ENSMUST00000170799.1 ENSMUST00000170422.2 |
Evi2a
Evi2b
|
ecotropic viral integration site 2a ecotropic viral integration site 2b |
| chrX_+_159697308 | 2.34 |
ENSMUST00000123433.1
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr14_+_70774304 | 2.25 |
ENSMUST00000022698.7
|
Dok2
|
docking protein 2 |
| chr18_-_62179948 | 2.23 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr6_-_136941694 | 2.23 |
ENSMUST00000032344.5
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta |
| chr2_+_153938218 | 2.23 |
ENSMUST00000109757.1
|
Bpifb4
|
BPI fold containing family B, member 4 |
| chr15_-_34356421 | 2.19 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr11_-_69895523 | 2.10 |
ENSMUST00000001631.6
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr2_-_119760295 | 2.08 |
ENSMUST00000082130.6
ENSMUST00000028759.6 |
Ltk
|
leukocyte tyrosine kinase |
| chr15_-_98607611 | 2.06 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr10_+_118441044 | 2.05 |
ENSMUST00000068592.3
|
Ifng
|
interferon gamma |
| chr2_-_73485733 | 2.00 |
ENSMUST00000102680.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr11_-_4594750 | 1.96 |
ENSMUST00000109943.3
|
Mtmr3
|
myotubularin related protein 3 |
| chr7_-_133709051 | 1.94 |
ENSMUST00000124759.1
ENSMUST00000106144.1 |
Uros
|
uroporphyrinogen III synthase |
| chr7_-_115824699 | 1.94 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr16_-_58718724 | 1.86 |
ENSMUST00000089318.3
|
Gpr15
|
G protein-coupled receptor 15 |
| chr4_-_129573637 | 1.81 |
ENSMUST00000102596.1
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr4_-_70410422 | 1.78 |
ENSMUST00000144099.1
|
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chr2_-_73486456 | 1.72 |
ENSMUST00000141264.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr7_-_25675047 | 1.69 |
ENSMUST00000108404.1
ENSMUST00000108405.1 ENSMUST00000079439.3 |
Tmem91
|
transmembrane protein 91 |
| chr18_+_62180119 | 1.68 |
ENSMUST00000067743.1
|
Gm9949
|
predicted gene 9949 |
| chr15_+_9436028 | 1.58 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr11_-_101095367 | 1.54 |
ENSMUST00000019447.8
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein |
| chr12_-_112860886 | 1.51 |
ENSMUST00000021729.7
|
Gpr132
|
G protein-coupled receptor 132 |
| chr5_+_139406387 | 1.48 |
ENSMUST00000052176.8
|
C130050O18Rik
|
RIKEN cDNA C130050O18 gene |
| chr17_+_75178797 | 1.48 |
ENSMUST00000112516.1
ENSMUST00000135447.1 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr3_-_96814518 | 1.46 |
ENSMUST00000047702.7
|
Cd160
|
CD160 antigen |
| chr15_-_97844254 | 1.44 |
ENSMUST00000119670.1
ENSMUST00000116409.2 |
Hdac7
|
histone deacetylase 7 |
| chr15_-_97844164 | 1.43 |
ENSMUST00000120683.1
|
Hdac7
|
histone deacetylase 7 |
| chr7_-_133708958 | 1.38 |
ENSMUST00000106146.1
|
Uros
|
uroporphyrinogen III synthase |
| chr7_+_126584937 | 1.34 |
ENSMUST00000039522.6
|
Apobr
|
apolipoprotein B receptor |
| chr17_+_75178911 | 1.29 |
ENSMUST00000112514.1
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr10_-_40302186 | 1.28 |
ENSMUST00000099945.4
|
Amd1
|
S-adenosylmethionine decarboxylase 1 |
| chr1_+_171388954 | 1.28 |
ENSMUST00000056449.8
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr7_-_133709069 | 1.27 |
ENSMUST00000106145.3
|
Uros
|
uroporphyrinogen III synthase |
| chr8_+_109705549 | 1.27 |
ENSMUST00000034163.8
|
Zfp821
|
zinc finger protein 821 |
| chr2_-_32741016 | 1.24 |
ENSMUST00000009695.2
|
6330409D20Rik
|
RIKEN cDNA 6330409D20 gene |
| chr2_+_153918391 | 1.24 |
ENSMUST00000109760.1
|
Bpifb3
|
BPI fold containing family B, member 3 |
| chr5_+_140607334 | 1.23 |
ENSMUST00000031555.1
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chrX_-_48034842 | 1.23 |
ENSMUST00000039026.7
|
Apln
|
apelin |
| chr8_+_94667082 | 1.20 |
ENSMUST00000109527.4
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr1_-_138175126 | 1.17 |
ENSMUST00000183301.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr19_+_5568002 | 1.16 |
ENSMUST00000096318.3
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_-_138175283 | 1.14 |
ENSMUST00000182755.1
ENSMUST00000183262.1 ENSMUST00000027645.7 ENSMUST00000112036.2 ENSMUST00000182283.1 |
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr5_+_48242549 | 1.12 |
ENSMUST00000172493.1
|
Slit2
|
slit homolog 2 (Drosophila) |
| chr9_-_32928928 | 1.11 |
ENSMUST00000185169.1
|
RP24-308I2.1
|
RP24-308I2.1 |
| chrX_+_56786527 | 1.10 |
ENSMUST00000144600.1
|
Fhl1
|
four and a half LIM domains 1 |
| chr1_-_138175238 | 1.09 |
ENSMUST00000182536.1
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C |
| chr4_-_41275091 | 1.09 |
ENSMUST00000030143.6
ENSMUST00000108068.1 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr2_+_172393794 | 1.07 |
ENSMUST00000099061.2
ENSMUST00000103073.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr3_-_86142684 | 1.04 |
ENSMUST00000029722.6
|
Rps3a1
|
ribosomal protein S3A1 |
| chr15_+_80711292 | 1.01 |
ENSMUST00000067689.7
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr16_-_95586585 | 0.93 |
ENSMUST00000077773.6
|
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chr16_+_18877037 | 0.91 |
ENSMUST00000120532.1
ENSMUST00000004222.7 |
Hira
|
histone cell cycle regulation defective homolog A (S. cerevisiae) |
| chr11_-_110168073 | 0.87 |
ENSMUST00000044850.3
|
Abca9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr10_-_95673451 | 0.87 |
ENSMUST00000099328.1
|
Anapc15-ps
|
anaphase prompoting complex C subunit 15, pseudogene |
| chr9_-_107770945 | 0.85 |
ENSMUST00000183248.1
ENSMUST00000182022.1 ENSMUST00000035199.6 ENSMUST00000182659.1 |
Rbm5
|
RNA binding motif protein 5 |
| chr11_-_100121558 | 0.83 |
ENSMUST00000007275.2
|
Krt13
|
keratin 13 |
| chr15_-_78493607 | 0.83 |
ENSMUST00000163494.1
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr4_-_6990774 | 0.83 |
ENSMUST00000039987.3
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr1_-_161788489 | 0.82 |
ENSMUST00000000834.2
|
Fasl
|
Fas ligand (TNF superfamily, member 6) |
| chr6_+_136518820 | 0.82 |
ENSMUST00000032335.6
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_+_133176336 | 0.75 |
ENSMUST00000105912.1
|
Wasf2
|
WAS protein family, member 2 |
| chr1_+_160044564 | 0.72 |
ENSMUST00000168359.1
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr4_-_152038568 | 0.71 |
ENSMUST00000030792.1
|
Tas1r1
|
taste receptor, type 1, member 1 |
| chrX_-_12160355 | 0.67 |
ENSMUST00000043441.6
|
Bcor
|
BCL6 interacting corepressor |
| chr1_-_164935522 | 0.59 |
ENSMUST00000027860.7
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr16_+_32271468 | 0.57 |
ENSMUST00000093183.3
|
Smco1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr6_-_38637220 | 0.55 |
ENSMUST00000096030.3
|
Klrg2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr10_-_127288999 | 0.54 |
ENSMUST00000119078.1
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr14_-_121915774 | 0.51 |
ENSMUST00000055475.7
|
Gpr18
|
G protein-coupled receptor 18 |
| chr1_+_172956768 | 0.48 |
ENSMUST00000038432.4
|
Olfr16
|
olfactory receptor 16 |
| chr9_+_113812547 | 0.45 |
ENSMUST00000166734.2
ENSMUST00000111838.2 ENSMUST00000163895.2 |
Clasp2
|
CLIP associating protein 2 |
| chr7_-_132852606 | 0.45 |
ENSMUST00000120425.1
|
Mettl10
|
methyltransferase like 10 |
| chr9_+_108479849 | 0.41 |
ENSMUST00000065014.4
|
Lamb2
|
laminin, beta 2 |
| chr12_+_116077720 | 0.41 |
ENSMUST00000011315.3
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr5_+_87808082 | 0.41 |
ENSMUST00000072539.5
ENSMUST00000113279.1 ENSMUST00000101057.3 |
Csn1s2b
|
casein alpha s2-like B |
| chr10_-_127288851 | 0.40 |
ENSMUST00000156208.1
ENSMUST00000026476.6 |
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr2_-_20943413 | 0.40 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr1_+_36307727 | 0.34 |
ENSMUST00000126413.1
|
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr2_+_37516618 | 0.33 |
ENSMUST00000065441.6
|
Gpr21
|
G protein-coupled receptor 21 |
| chr11_+_75531690 | 0.30 |
ENSMUST00000149727.1
ENSMUST00000042561.7 ENSMUST00000108433.1 ENSMUST00000143035.1 |
Slc43a2
|
solute carrier family 43, member 2 |
| chrX_-_20950597 | 0.29 |
ENSMUST00000009550.7
|
Elk1
|
ELK1, member of ETS oncogene family |
| chr9_-_66514567 | 0.28 |
ENSMUST00000056890.8
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr19_+_4214238 | 0.26 |
ENSMUST00000046506.6
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr12_-_34528844 | 0.26 |
ENSMUST00000110819.2
|
Hdac9
|
histone deacetylase 9 |
| chr14_+_56017964 | 0.24 |
ENSMUST00000022836.4
|
Mcpt1
|
mast cell protease 1 |
| chr10_-_14718191 | 0.22 |
ENSMUST00000020016.4
|
Gje1
|
gap junction protein, epsilon 1 |
| chr15_+_62037986 | 0.22 |
ENSMUST00000182956.1
ENSMUST00000182075.1 ENSMUST00000180432.2 ENSMUST00000181416.2 ENSMUST00000181657.2 |
Pvt1
|
plasmacytoma variant translocation 1 |
| chr2_+_86041317 | 0.21 |
ENSMUST00000111589.1
|
Olfr1033
|
olfactory receptor 1033 |
| chr2_-_71367749 | 0.20 |
ENSMUST00000151937.1
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr3_-_88378699 | 0.19 |
ENSMUST00000098956.2
|
Bglap2
|
bone gamma-carboxyglutamate protein 2 |
| chr1_-_174118014 | 0.18 |
ENSMUST00000063030.3
|
Olfr231
|
olfactory receptor 231 |
| chr16_+_91391721 | 0.18 |
ENSMUST00000160764.1
|
Gm21970
|
predicted gene 21970 |
| chr11_+_77686155 | 0.18 |
ENSMUST00000100802.4
ENSMUST00000181023.1 |
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr17_-_44814649 | 0.18 |
ENSMUST00000113571.3
|
Runx2
|
runt related transcription factor 2 |
| chr16_+_25801907 | 0.16 |
ENSMUST00000040231.6
ENSMUST00000115306.1 ENSMUST00000115304.1 ENSMUST00000115305.1 |
Trp63
|
transformation related protein 63 |
| chr11_+_75532127 | 0.15 |
ENSMUST00000127226.1
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr19_-_8839181 | 0.15 |
ENSMUST00000096259.4
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr17_-_44814604 | 0.15 |
ENSMUST00000160673.1
|
Runx2
|
runt related transcription factor 2 |
| chr16_-_38433145 | 0.15 |
ENSMUST00000002926.6
|
Pla1a
|
phospholipase A1 member A |
| chr1_+_60746358 | 0.14 |
ENSMUST00000027165.2
|
Cd28
|
CD28 antigen |
| chr2_+_35691893 | 0.13 |
ENSMUST00000065001.5
|
Dab2ip
|
disabled 2 interacting protein |
| chr15_+_62178175 | 0.11 |
ENSMUST00000182476.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr1_+_36307745 | 0.10 |
ENSMUST00000142319.1
ENSMUST00000097778.2 ENSMUST00000115031.1 ENSMUST00000115032.1 ENSMUST00000137906.1 ENSMUST00000115029.1 |
Arid5a
|
AT rich interactive domain 5A (MRF1-like) |
| chr6_-_83070225 | 0.08 |
ENSMUST00000174674.1
ENSMUST00000089641.3 |
Tlx2
|
T cell leukemia, homeobox 2 |
| chr12_+_95692212 | 0.08 |
ENSMUST00000057324.3
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr3_-_107760221 | 0.08 |
ENSMUST00000153114.1
ENSMUST00000118593.1 ENSMUST00000120243.1 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr14_-_20618339 | 0.08 |
ENSMUST00000035340.7
|
Usp54
|
ubiquitin specific peptidase 54 |
| chr7_-_132852657 | 0.07 |
ENSMUST00000033257.8
|
Mettl10
|
methyltransferase like 10 |
| chr19_+_5050807 | 0.06 |
ENSMUST00000025818.6
|
Rin1
|
Ras and Rab interactor 1 |
| chr4_+_102991784 | 0.06 |
ENSMUST00000140654.1
ENSMUST00000169211.1 |
Tctex1d1
|
Tctex1 domain containing 1 |
| chr15_-_38519227 | 0.05 |
ENSMUST00000110328.1
ENSMUST00000110329.1 ENSMUST00000065308.5 |
Azin1
|
antizyme inhibitor 1 |
| chrX_-_50942710 | 0.05 |
ENSMUST00000060650.5
|
Frmd7
|
FERM domain containing 7 |
| chr4_+_65124174 | 0.05 |
ENSMUST00000084501.3
|
Pappa
|
pregnancy-associated plasma protein A |
| chr18_+_37411674 | 0.04 |
ENSMUST00000051126.2
|
Pcdhb10
|
protocadherin beta 10 |
| chr11_-_72215592 | 0.04 |
ENSMUST00000021157.8
|
Med31
|
mediator of RNA polymerase II transcription, subunit 31 homolog (yeast) |
| chr2_+_32629467 | 0.04 |
ENSMUST00000068271.4
|
Ak1
|
adenylate kinase 1 |
| chr15_-_102722120 | 0.03 |
ENSMUST00000171838.1
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr11_-_102218923 | 0.02 |
ENSMUST00000131254.1
|
Hdac5
|
histone deacetylase 5 |
| chr17_-_36958533 | 0.02 |
ENSMUST00000172518.1
|
Znrd1
|
zinc ribbon domain containing, 1 |
| chr15_-_102722150 | 0.02 |
ENSMUST00000023818.3
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr3_+_103058302 | 0.01 |
ENSMUST00000029445.6
|
Nras
|
neuroblastoma ras oncogene |
| chr11_-_52282564 | 0.00 |
ENSMUST00000086844.3
|
Tcf7
|
transcription factor 7, T cell specific |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 29.7 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 4.4 | 13.2 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 2.7 | 10.6 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 2.3 | 6.8 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.9 | 5.8 | GO:0002865 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) negative regulation of hypersensitivity(GO:0002884) |
| 1.5 | 7.7 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 1.5 | 4.5 | GO:0071663 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 1.4 | 6.9 | GO:0070103 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 1.1 | 4.6 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.7 | 4.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.7 | 6.4 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.7 | 8.5 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.7 | 2.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.6 | 6.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.6 | 4.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.6 | 2.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.6 | 2.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.5 | 1.5 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.5 | 2.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.4 | 1.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.4 | 5.5 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.4 | 1.1 | GO:0021972 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.3 | 2.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.3 | 3.0 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 1.8 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 3.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 5.0 | GO:0038065 | collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 2.5 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.2 | 1.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.2 | 2.7 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.2 | 0.4 | GO:2000554 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.2 | 3.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 0.4 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.2 | 3.6 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.2 | 3.0 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 2.5 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.2 | 1.8 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.2 | 1.9 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 | 0.5 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 0.7 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.2 | 0.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.2 | 0.2 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.2 | 2.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.2 | 0.9 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 3.9 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 2.7 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.7 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 3.7 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 2.8 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.1 | 2.8 | GO:0010714 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.1 | 1.0 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.4 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.4 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 2.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 2.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 9.1 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 5.4 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 1.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.9 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 2.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:1903972 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 1.5 | GO:0035825 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 1.5 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 1.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 1.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0046641 | positive regulation of alpha-beta T cell proliferation(GO:0046641) |
| 0.0 | 3.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 29.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.5 | 6.0 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.9 | 2.8 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.8 | 10.6 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.7 | 2.9 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.5 | 5.6 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.4 | 5.8 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 3.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.4 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 13.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 1.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) chylomicron(GO:0042627) |
| 0.1 | 0.3 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 1.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 6.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 8.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 3.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 5.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 1.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 4.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 4.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 5.1 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.2 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 4.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 2.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.2 | 10.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.8 | 4.6 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.7 | 2.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 7.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.6 | 4.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.6 | 2.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.5 | 7.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.5 | 29.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.4 | 1.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.3 | 2.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.3 | 13.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 0.8 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 2.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.3 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.2 | 5.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 1.3 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.2 | 8.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 3.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 5.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 3.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0043237 | GTPase inhibitor activity(GO:0005095) laminin-1 binding(GO:0043237) Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 3.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 3.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 3.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 2.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.3 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 0.7 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.1 | 3.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 4.3 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.1 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 8.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 1.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 6.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.1 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 3.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.0 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 2.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 34.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.5 | 5.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.3 | 16.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 14.7 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 6.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 6.4 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 7.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 5.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 3.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 4.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 6.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 4.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 4.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 5.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 5.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 2.0 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 2.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 2.7 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 3.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 3.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 3.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 2.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 8.5 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 2.2 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 1.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 3.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 2.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 4.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 2.1 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |