Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Rreb1
Z-value: 1.13
Transcription factors associated with Rreb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rreb1
|
ENSMUSG00000039087.10 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rreb1 | mm10_v2_chr13_+_37778384_37778403 | 0.53 | 8.3e-04 | Click! |
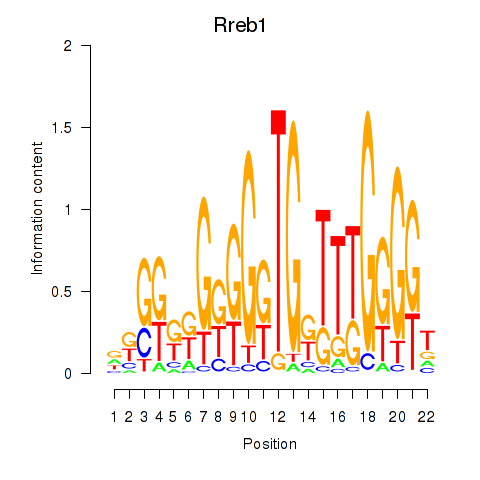
Activity profile of Rreb1 motif
Sorted Z-values of Rreb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_142576492 | 5.74 |
ENSMUST00000140716.1
|
H19
|
H19 fetal liver mRNA |
| chr16_-_36408349 | 5.52 |
ENSMUST00000023619.6
|
Stfa2
|
stefin A2 |
| chr9_-_109849440 | 4.19 |
ENSMUST00000112022.2
|
Camp
|
cathelicidin antimicrobial peptide |
| chr7_-_142578093 | 3.17 |
ENSMUST00000149974.1
ENSMUST00000152754.1 |
H19
|
H19 fetal liver mRNA |
| chr7_-_142578139 | 3.14 |
ENSMUST00000136359.1
|
H19
|
H19 fetal liver mRNA |
| chr5_-_122002340 | 2.67 |
ENSMUST00000134326.1
|
Cux2
|
cut-like homeobox 2 |
| chr17_+_29135056 | 2.28 |
ENSMUST00000087942.4
|
Rab44
|
RAB44, member RAS oncogene family |
| chr4_+_44943727 | 2.04 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr8_+_57455898 | 1.77 |
ENSMUST00000034023.3
|
Scrg1
|
scrapie responsive gene 1 |
| chr14_+_32321987 | 1.75 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr12_+_109545390 | 1.74 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr9_-_103480328 | 1.73 |
ENSMUST00000124310.2
|
Bfsp2
|
beaded filament structural protein 2, phakinin |
| chr6_+_127887582 | 1.70 |
ENSMUST00000032501.4
|
Tspan11
|
tetraspanin 11 |
| chr7_+_24897381 | 1.63 |
ENSMUST00000003469.7
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chrX_-_73659724 | 1.52 |
ENSMUST00000114473.1
ENSMUST00000002087.7 |
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase |
| chr4_-_4138432 | 1.51 |
ENSMUST00000070375.7
|
Penk
|
preproenkephalin |
| chr5_+_76656512 | 1.49 |
ENSMUST00000086909.4
|
Gm10430
|
predicted gene 10430 |
| chr14_+_69609068 | 1.41 |
ENSMUST00000022660.7
|
Loxl2
|
lysyl oxidase-like 2 |
| chr6_+_72549652 | 1.40 |
ENSMUST00000134809.1
|
Capg
|
capping protein (actin filament), gelsolin-like |
| chr4_+_115059507 | 1.32 |
ENSMUST00000162489.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr14_+_55765956 | 1.30 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr4_+_41633286 | 1.30 |
ENSMUST00000119127.1
|
Dnaic1
|
dynein, axonemal, intermediate chain 1 |
| chr3_+_96181151 | 1.27 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr2_-_158229215 | 1.27 |
ENSMUST00000103121.3
ENSMUST00000169335.1 ENSMUST00000046944.5 |
D630003M21Rik
|
RIKEN cDNA D630003M21 gene |
| chr9_-_67832325 | 1.26 |
ENSMUST00000054500.5
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr4_-_3938354 | 1.23 |
ENSMUST00000003369.3
|
Plag1
|
pleiomorphic adenoma gene 1 |
| chr19_+_8591254 | 1.22 |
ENSMUST00000010251.3
ENSMUST00000170817.1 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr6_+_87778084 | 1.21 |
ENSMUST00000032133.3
|
Gp9
|
glycoprotein 9 (platelet) |
| chr8_-_11008458 | 1.18 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr5_+_122210134 | 1.16 |
ENSMUST00000100747.2
|
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr4_+_44300876 | 1.10 |
ENSMUST00000045607.5
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr11_-_106314494 | 1.07 |
ENSMUST00000167143.1
|
Cd79b
|
CD79B antigen |
| chr11_-_94242701 | 1.04 |
ENSMUST00000061469.3
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr11_-_100411874 | 1.03 |
ENSMUST00000141840.1
|
Leprel4
|
leprecan-like 4 |
| chr7_+_100495987 | 1.03 |
ENSMUST00000133044.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chrX_+_7919816 | 1.01 |
ENSMUST00000041096.3
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr5_+_135674572 | 1.00 |
ENSMUST00000153515.1
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr17_+_43568096 | 0.97 |
ENSMUST00000167214.1
|
Pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr1_+_187608755 | 0.97 |
ENSMUST00000127489.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr3_+_146149829 | 0.96 |
ENSMUST00000011152.7
|
Mcoln2
|
mucolipin 2 |
| chr4_+_136172367 | 0.94 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr13_+_76579670 | 0.94 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr8_-_95294074 | 0.92 |
ENSMUST00000184103.1
|
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr8_+_120002720 | 0.89 |
ENSMUST00000108972.3
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr11_-_103356324 | 0.89 |
ENSMUST00000136491.2
ENSMUST00000107023.2 |
Arhgap27
|
Rho GTPase activating protein 27 |
| chr6_+_145121727 | 0.88 |
ENSMUST00000032396.6
|
Lrmp
|
lymphoid-restricted membrane protein |
| chr10_+_78069351 | 0.88 |
ENSMUST00000105393.1
|
Icosl
|
icos ligand |
| chr3_+_146150174 | 0.88 |
ENSMUST00000098524.4
|
Mcoln2
|
mucolipin 2 |
| chr3_+_51224447 | 0.87 |
ENSMUST00000023849.8
ENSMUST00000167780.1 |
Ccrn4l
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
| chr3_-_89245159 | 0.86 |
ENSMUST00000090924.6
|
Trim46
|
tripartite motif-containing 46 |
| chr7_+_5056856 | 0.84 |
ENSMUST00000131368.1
ENSMUST00000123956.1 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr12_-_112802646 | 0.80 |
ENSMUST00000124526.1
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr18_-_21652362 | 0.80 |
ENSMUST00000049105.4
|
Klhl14
|
kelch-like 14 |
| chr5_+_37047464 | 0.80 |
ENSMUST00000137019.1
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr9_+_119052863 | 0.78 |
ENSMUST00000131647.1
|
Vill
|
villin-like |
| chr19_-_4191035 | 0.78 |
ENSMUST00000045864.2
|
Tbc1d10c
|
TBC1 domain family, member 10c |
| chr4_-_11965699 | 0.78 |
ENSMUST00000108301.1
ENSMUST00000095144.3 ENSMUST00000108302.1 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chrX_-_73149450 | 0.78 |
ENSMUST00000080324.3
|
DXBay18
|
DNA segment, Chr X, Baylor 18 |
| chr10_-_6980376 | 0.77 |
ENSMUST00000105617.1
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chrX_-_104671048 | 0.77 |
ENSMUST00000042070.5
|
Zdhhc15
|
zinc finger, DHHC domain containing 15 |
| chr7_-_133123770 | 0.75 |
ENSMUST00000164896.1
ENSMUST00000171968.1 |
Ctbp2
|
C-terminal binding protein 2 |
| chr5_+_122100951 | 0.75 |
ENSMUST00000014080.6
ENSMUST00000111750.1 ENSMUST00000139213.1 ENSMUST00000111751.1 ENSMUST00000155612.1 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr19_-_6858179 | 0.75 |
ENSMUST00000113440.1
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr1_-_182019927 | 0.75 |
ENSMUST00000078719.6
ENSMUST00000111030.3 ENSMUST00000177811.1 ENSMUST00000111024.3 ENSMUST00000111025.2 |
Enah
|
enabled homolog (Drosophila) |
| chr11_-_96005872 | 0.74 |
ENSMUST00000013559.2
|
Igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr13_+_52596847 | 0.74 |
ENSMUST00000055087.6
|
Syk
|
spleen tyrosine kinase |
| chr2_-_125506385 | 0.74 |
ENSMUST00000028633.6
|
Fbn1
|
fibrillin 1 |
| chrX_+_73117047 | 0.73 |
ENSMUST00000088459.2
|
Gm14685
|
predicted gene 14685 |
| chr4_+_62480025 | 0.71 |
ENSMUST00000030088.5
ENSMUST00000107449.3 |
Bspry
|
B-box and SPRY domain containing |
| chr11_+_3289880 | 0.69 |
ENSMUST00000110043.1
ENSMUST00000094471.3 |
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_+_25627604 | 0.69 |
ENSMUST00000076034.6
|
B3gnt8
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
| chr11_-_59029509 | 0.69 |
ENSMUST00000127937.1
|
Obscn
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF |
| chr17_+_47593516 | 0.69 |
ENSMUST00000182874.1
|
Ccnd3
|
cyclin D3 |
| chr2_+_158306493 | 0.69 |
ENSMUST00000016168.2
ENSMUST00000109491.1 |
Lbp
|
lipopolysaccharide binding protein |
| chr2_+_145167706 | 0.69 |
ENSMUST00000110007.1
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr16_-_32165454 | 0.69 |
ENSMUST00000115163.3
ENSMUST00000144345.1 ENSMUST00000143682.1 ENSMUST00000115165.3 ENSMUST00000099991.4 ENSMUST00000130410.1 |
Nrros
|
negative regulator of reactive oxygen species |
| chr2_+_136057927 | 0.68 |
ENSMUST00000057503.6
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr10_-_7473361 | 0.68 |
ENSMUST00000169796.2
ENSMUST00000177585.1 |
Ulbp1
|
UL16 binding protein 1 |
| chr6_+_48860339 | 0.68 |
ENSMUST00000101425.3
|
Gm7932
|
predicted gene 7932 |
| chr1_+_135783065 | 0.67 |
ENSMUST00000132795.1
|
Tnni1
|
troponin I, skeletal, slow 1 |
| chr9_+_119052770 | 0.67 |
ENSMUST00000051386.6
ENSMUST00000074734.6 |
Vill
|
villin-like |
| chr4_+_130915949 | 0.67 |
ENSMUST00000030316.6
|
Laptm5
|
lysosomal-associated protein transmembrane 5 |
| chr9_+_110333402 | 0.67 |
ENSMUST00000133114.1
ENSMUST00000125759.1 |
Scap
|
SREBF chaperone |
| chr19_-_56389877 | 0.66 |
ENSMUST00000166203.1
ENSMUST00000167239.1 ENSMUST00000040711.8 ENSMUST00000095947.4 ENSMUST00000073536.6 |
Nrap
|
nebulin-related anchoring protein |
| chr18_-_68429235 | 0.65 |
ENSMUST00000052347.6
|
Mc2r
|
melanocortin 2 receptor |
| chr5_+_113490447 | 0.65 |
ENSMUST00000094452.3
|
Wscd2
|
WSC domain containing 2 |
| chr2_+_103957976 | 0.64 |
ENSMUST00000156813.1
ENSMUST00000170926.1 |
Lmo2
|
LIM domain only 2 |
| chr15_-_78529617 | 0.64 |
ENSMUST00000023075.8
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr8_+_25911670 | 0.64 |
ENSMUST00000120653.1
ENSMUST00000126226.1 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr15_-_78855517 | 0.63 |
ENSMUST00000044584.4
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr14_+_70077375 | 0.63 |
ENSMUST00000035908.1
|
Egr3
|
early growth response 3 |
| chr6_+_72549430 | 0.62 |
ENSMUST00000155705.1
|
Capg
|
capping protein (actin filament), gelsolin-like |
| chr7_+_24862193 | 0.62 |
ENSMUST00000052897.4
ENSMUST00000170837.2 |
Gm9844
Gm9844
|
predicted pseudogene 9844 predicted pseudogene 9844 |
| chr17_+_37002610 | 0.61 |
ENSMUST00000173921.1
ENSMUST00000172580.1 |
Zfp57
|
zinc finger protein 57 |
| chr3_+_88621102 | 0.60 |
ENSMUST00000029694.7
ENSMUST00000176804.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr9_-_32542861 | 0.60 |
ENSMUST00000183767.1
|
Fli1
|
Friend leukemia integration 1 |
| chr1_-_72874877 | 0.60 |
ENSMUST00000027377.8
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr3_-_89245005 | 0.59 |
ENSMUST00000107464.1
|
Trim46
|
tripartite motif-containing 46 |
| chr5_+_122209729 | 0.59 |
ENSMUST00000072602.7
ENSMUST00000143560.1 |
Hvcn1
|
hydrogen voltage-gated channel 1 |
| chr18_-_47333311 | 0.58 |
ENSMUST00000126684.1
ENSMUST00000156422.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr15_+_80133114 | 0.58 |
ENSMUST00000023050.7
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr7_-_126594941 | 0.58 |
ENSMUST00000058429.5
|
Il27
|
interleukin 27 |
| chr19_-_4283033 | 0.58 |
ENSMUST00000167215.1
ENSMUST00000056888.6 |
Ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr6_+_141249161 | 0.57 |
ENSMUST00000043259.7
|
Pde3a
|
phosphodiesterase 3A, cGMP inhibited |
| chr5_-_111761697 | 0.57 |
ENSMUST00000129146.1
ENSMUST00000137398.1 ENSMUST00000129065.1 |
E130006D01Rik
|
RIKEN cDNA E130006D01 gene |
| chr18_+_36679575 | 0.56 |
ENSMUST00000050476.4
ENSMUST00000036158.6 |
Slc35a4
|
solute carrier family 35, member A4 |
| chr17_+_47593444 | 0.56 |
ENSMUST00000182209.1
|
Ccnd3
|
cyclin D3 |
| chrX_-_57338598 | 0.56 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr14_-_34201604 | 0.55 |
ENSMUST00000096019.2
|
Gprin2
|
G protein regulated inducer of neurite outgrowth 2 |
| chr2_+_147364989 | 0.55 |
ENSMUST00000109968.2
|
Pax1
|
paired box gene 1 |
| chr7_+_43781054 | 0.55 |
ENSMUST00000014058.9
|
Klk10
|
kallikrein related-peptidase 10 |
| chr9_+_110333276 | 0.54 |
ENSMUST00000125823.1
ENSMUST00000131328.1 |
Scap
|
SREBF chaperone |
| chr6_-_126698192 | 0.54 |
ENSMUST00000060972.3
|
Kcna5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chr15_-_74636241 | 0.53 |
ENSMUST00000023271.1
|
Mroh4
|
maestro heat-like repeat family member 4 |
| chr18_+_46850034 | 0.53 |
ENSMUST00000025358.2
|
4833403I15Rik
|
RIKEN cDNA 4833403I15 gene |
| chr11_-_114960417 | 0.53 |
ENSMUST00000092466.5
ENSMUST00000061637.3 |
Cd300c
|
CD300C antigen |
| chr13_-_98206151 | 0.53 |
ENSMUST00000109426.1
|
Arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr11_+_77765588 | 0.53 |
ENSMUST00000164315.1
|
Myo18a
|
myosin XVIIIA |
| chrX_-_48034842 | 0.52 |
ENSMUST00000039026.7
|
Apln
|
apelin |
| chr5_+_140735526 | 0.51 |
ENSMUST00000120630.2
|
Amz1
|
archaelysin family metallopeptidase 1 |
| chr6_+_72549252 | 0.51 |
ENSMUST00000114071.1
|
Capg
|
capping protein (actin filament), gelsolin-like |
| chr6_+_89140992 | 0.51 |
ENSMUST00000162900.1
ENSMUST00000161362.1 ENSMUST00000160361.1 ENSMUST00000101176.3 |
Gm1965
Gm1965
|
predicted gene 1965 predicted gene 1965 |
| chr8_+_69808672 | 0.50 |
ENSMUST00000036074.8
ENSMUST00000123453.1 |
Gmip
|
Gem-interacting protein |
| chr11_-_34208085 | 0.50 |
ENSMUST00000060271.2
|
Foxi1
|
forkhead box I1 |
| chr6_+_29396665 | 0.49 |
ENSMUST00000096084.5
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr17_+_35077080 | 0.49 |
ENSMUST00000172959.1
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr6_+_17307040 | 0.49 |
ENSMUST00000123439.1
|
Cav1
|
caveolin 1, caveolae protein |
| chr4_-_134012381 | 0.49 |
ENSMUST00000176113.1
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_14873656 | 0.48 |
ENSMUST00000114718.1
ENSMUST00000114719.1 |
Cacnb2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr5_-_89883321 | 0.48 |
ENSMUST00000163159.1
ENSMUST00000061427.5 |
Adamts3
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 3 |
| chr9_-_8134294 | 0.47 |
ENSMUST00000037397.6
|
AK129341
|
cDNA sequence AK129341 |
| chr1_-_37719782 | 0.47 |
ENSMUST00000160589.1
|
2010300C02Rik
|
RIKEN cDNA 2010300C02 gene |
| chr7_+_90426312 | 0.46 |
ENSMUST00000061391.7
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr17_+_37002574 | 0.46 |
ENSMUST00000173588.1
|
Zfp57
|
zinc finger protein 57 |
| chr4_-_58206596 | 0.46 |
ENSMUST00000042850.8
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr7_-_6155939 | 0.45 |
ENSMUST00000094870.1
|
Zfp787
|
zinc finger protein 787 |
| chr8_+_57320975 | 0.45 |
ENSMUST00000040104.3
|
Hand2
|
heart and neural crest derivatives expressed transcript 2 |
| chr7_+_127777095 | 0.44 |
ENSMUST00000144406.1
|
Setd1a
|
SET domain containing 1A |
| chr11_-_98983016 | 0.44 |
ENSMUST00000062931.1
|
Gjd3
|
gap junction protein, delta 3 |
| chr11_+_116198853 | 0.43 |
ENSMUST00000021130.6
|
Ten1
|
TEN1 telomerase capping complex subunit |
| chr3_-_32616479 | 0.43 |
ENSMUST00000108234.1
ENSMUST00000155737.1 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr3_-_89245829 | 0.42 |
ENSMUST00000041022.8
|
Trim46
|
tripartite motif-containing 46 |
| chr2_-_45113255 | 0.42 |
ENSMUST00000068415.4
ENSMUST00000127520.1 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr17_+_37002510 | 0.42 |
ENSMUST00000069250.7
|
Zfp57
|
zinc finger protein 57 |
| chr7_-_98178254 | 0.42 |
ENSMUST00000040971.7
|
Capn5
|
calpain 5 |
| chr7_+_73391160 | 0.42 |
ENSMUST00000128471.1
|
Rgma
|
RGM domain family, member A |
| chr19_+_10525244 | 0.42 |
ENSMUST00000038379.3
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr2_-_163417092 | 0.42 |
ENSMUST00000127038.1
|
Oser1
|
oxidative stress responsive serine rich 1 |
| chr19_+_38395980 | 0.41 |
ENSMUST00000054098.2
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr2_-_9878580 | 0.41 |
ENSMUST00000102976.3
|
Gata3
|
GATA binding protein 3 |
| chr5_+_135994796 | 0.41 |
ENSMUST00000111142.2
ENSMUST00000111145.3 ENSMUST00000111144.1 ENSMUST00000005072.3 ENSMUST00000130345.1 |
Dtx2
|
deltex 2 homolog (Drosophila) |
| chr2_-_31116289 | 0.41 |
ENSMUST00000149196.1
|
Fnbp1
|
formin binding protein 1 |
| chr6_-_92481343 | 0.41 |
ENSMUST00000113445.1
|
Prickle2
|
prickle homolog 2 (Drosophila) |
| chr6_+_29396576 | 0.40 |
ENSMUST00000115275.1
|
Ccdc136
|
coiled-coil domain containing 136 |
| chr1_+_171214013 | 0.40 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr3_+_159839729 | 0.40 |
ENSMUST00000068952.5
|
Wls
|
wntless homolog (Drosophila) |
| chr9_-_66919646 | 0.39 |
ENSMUST00000041139.7
|
Rab8b
|
RAB8B, member RAS oncogene family |
| chr5_-_73632421 | 0.39 |
ENSMUST00000087177.2
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr17_+_37002482 | 0.39 |
ENSMUST00000172527.1
|
Zfp57
|
zinc finger protein 57 |
| chr3_-_125938537 | 0.39 |
ENSMUST00000057944.7
|
Ugt8a
|
UDP galactosyltransferase 8A |
| chr4_-_141598206 | 0.39 |
ENSMUST00000131317.1
ENSMUST00000006381.4 ENSMUST00000129602.1 |
Fblim1
|
filamin binding LIM protein 1 |
| chr3_-_96594128 | 0.38 |
ENSMUST00000145001.1
ENSMUST00000091924.3 |
Polr3gl
|
polymerase (RNA) III (DNA directed) polypeptide G like |
| chr14_+_70555900 | 0.38 |
ENSMUST00000163060.1
|
Hr
|
hairless |
| chr5_+_114707760 | 0.38 |
ENSMUST00000094441.4
|
Tchp
|
trichoplein, keratin filament binding |
| chr7_+_139214661 | 0.38 |
ENSMUST00000135509.1
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr11_+_121204420 | 0.38 |
ENSMUST00000038831.8
ENSMUST00000106117.1 |
Hexdc
|
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
| chr6_+_118066356 | 0.37 |
ENSMUST00000164960.1
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr5_+_34999111 | 0.37 |
ENSMUST00000114283.1
|
Rgs12
|
regulator of G-protein signaling 12 |
| chr2_+_74681991 | 0.37 |
ENSMUST00000142312.1
|
Hoxd11
|
homeobox D11 |
| chr19_+_24673998 | 0.37 |
ENSMUST00000057243.4
|
Tmem252
|
transmembrane protein 252 |
| chr3_-_88621061 | 0.37 |
ENSMUST00000177314.1
|
Gm20652
|
predicted gene 20652 |
| chrX_-_51205773 | 0.37 |
ENSMUST00000114875.1
|
Mbnl3
|
muscleblind-like 3 (Drosophila) |
| chr14_+_65806066 | 0.36 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr7_+_5056706 | 0.36 |
ENSMUST00000144802.1
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr8_+_93810832 | 0.36 |
ENSMUST00000034198.8
ENSMUST00000125716.1 |
Gnao1
|
guanine nucleotide binding protein, alpha O |
| chr4_-_41098174 | 0.36 |
ENSMUST00000055327.7
|
Aqp3
|
aquaporin 3 |
| chr6_-_89362581 | 0.36 |
ENSMUST00000163139.1
|
Plxna1
|
plexin A1 |
| chr10_+_68723723 | 0.35 |
ENSMUST00000080995.6
|
Tmem26
|
transmembrane protein 26 |
| chr9_-_37147257 | 0.35 |
ENSMUST00000039674.5
ENSMUST00000080754.5 |
Pknox2
|
Pbx/knotted 1 homeobox 2 |
| chr19_+_27217011 | 0.35 |
ENSMUST00000164746.1
ENSMUST00000172302.1 |
Vldlr
|
very low density lipoprotein receptor |
| chr19_+_27217357 | 0.35 |
ENSMUST00000047645.6
ENSMUST00000167487.1 |
Vldlr
|
very low density lipoprotein receptor |
| chr4_+_43441939 | 0.35 |
ENSMUST00000060864.6
|
Tesk1
|
testis specific protein kinase 1 |
| chr17_+_29549783 | 0.34 |
ENSMUST00000048677.7
|
Tbc1d22b
|
TBC1 domain family, member 22B |
| chr5_-_120588613 | 0.34 |
ENSMUST00000046426.8
|
Tpcn1
|
two pore channel 1 |
| chr4_-_117929726 | 0.34 |
ENSMUST00000070816.2
|
Artn
|
artemin |
| chr8_+_15057646 | 0.34 |
ENSMUST00000033842.3
|
Myom2
|
myomesin 2 |
| chr10_+_80179482 | 0.34 |
ENSMUST00000003154.6
|
Efna2
|
ephrin A2 |
| chr17_+_35823509 | 0.34 |
ENSMUST00000173493.1
ENSMUST00000173147.1 ENSMUST00000172846.1 |
Flot1
|
flotillin 1 |
| chr7_+_131548755 | 0.34 |
ENSMUST00000183219.1
|
Hmx2
|
H6 homeobox 2 |
| chr11_-_70229677 | 0.33 |
ENSMUST00000153449.1
ENSMUST00000000326.5 |
Bcl6b
|
B cell CLL/lymphoma 6, member B |
| chr9_-_90255927 | 0.33 |
ENSMUST00000144646.1
|
Tbc1d2b
|
TBC1 domain family, member 2B |
| chr11_-_50827681 | 0.33 |
ENSMUST00000109135.2
|
Zfp354c
|
zinc finger protein 354C |
| chr18_+_42053648 | 0.33 |
ENSMUST00000072008.4
|
Sh3rf2
|
SH3 domain containing ring finger 2 |
| chr8_-_87959560 | 0.33 |
ENSMUST00000109655.2
|
Zfp423
|
zinc finger protein 423 |
| chr6_+_48739039 | 0.33 |
ENSMUST00000054368.4
ENSMUST00000140054.1 |
Gimap1
|
GTPase, IMAP family member 1 |
| chr19_+_21778325 | 0.32 |
ENSMUST00000096194.2
ENSMUST00000025663.6 |
Tmem2
|
transmembrane protein 2 |
| chr5_-_114690906 | 0.32 |
ENSMUST00000112212.1
ENSMUST00000112214.1 |
Gltp
|
glycolipid transfer protein |
| chr2_-_65567465 | 0.32 |
ENSMUST00000066432.5
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr16_+_90826719 | 0.32 |
ENSMUST00000099548.2
|
Eva1c
|
eva-1 homolog C (C. elegans) |
| chr13_+_37825975 | 0.31 |
ENSMUST00000138043.1
|
Rreb1
|
ras responsive element binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rreb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 12.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.4 | 1.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.4 | 1.2 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 | 0.9 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 1.5 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.3 | 4.3 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.2 | 1.0 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.2 | 0.7 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.2 | 0.4 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.2 | 0.7 | GO:0045425 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.2 | 0.7 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 0.5 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.2 | 0.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.2 | 0.8 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 1.0 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.2 | 0.8 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.2 | 0.5 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.2 | 1.7 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.6 | GO:1904049 | negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.1 | 0.7 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 2.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.1 | 0.9 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.5 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.1 | 1.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.4 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.3 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.3 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 1.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 2.3 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.1 | 0.7 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 1.2 | GO:0010748 | negative regulation of B cell apoptotic process(GO:0002903) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 0.7 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.4 | GO:0090095 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.3 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.4 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.6 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.1 | 0.5 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 2.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.5 | GO:0098914 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.6 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.3 | GO:0061153 | negative regulation of interleukin-13 production(GO:0032696) trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) odontoblast differentiation(GO:0071895) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 0.5 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.3 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 1.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 2.0 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 3.0 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.1 | 1.0 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 1.4 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.2 | GO:0060857 | establishment of glial blood-brain barrier(GO:0060857) |
| 0.1 | 1.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.1 | 1.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.6 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.1 | 0.5 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.5 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.2 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.3 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.1 | 0.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.9 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 1.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0072137 | cell migration involved in endocardial cushion formation(GO:0003273) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:1902617 | response to fluoride(GO:1902617) |
| 0.1 | 0.3 | GO:0015867 | ATP transport(GO:0015867) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.6 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.2 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.0 | 0.6 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.5 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 2.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 1.0 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.8 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 | 0.8 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.2 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 1.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 3.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.1 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.4 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.2 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.0 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.4 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.2 | GO:1900242 | regulation of synaptic vesicle endocytosis(GO:1900242) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.2 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.9 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0030819 | positive regulation of cAMP biosynthetic process(GO:0030819) |
| 0.0 | 0.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 1.7 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 1.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.4 | 1.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 1.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 4.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.2 | 2.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.2 | 0.5 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 1.8 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 10.6 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.8 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.1 | 0.6 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 1.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.7 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.8 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.7 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.0 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.4 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 2.1 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0045179 | apical cortex(GO:0045179) ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:1990462 | omegasome(GO:1990462) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.8 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.9 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 5.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.7 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.3 | 1.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.3 | 1.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.3 | 1.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 1.0 | GO:0008941 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.2 | 0.7 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 0.2 | 0.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 1.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.8 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.2 | 0.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.5 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 1.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.8 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.2 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.1 | 0.7 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.7 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 1.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.4 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 0.6 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.1 | 0.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 1.0 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.5 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 4.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.2 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 1.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.2 | GO:0004063 | aryldialkylphosphatase activity(GO:0004063) |
| 0.0 | 2.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.7 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.5 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.5 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.6 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0008147 | structural constituent of bone(GO:0008147) hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0005346 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0051861 | glycolipid binding(GO:0051861) |
| 0.0 | 4.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 2.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 3.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.9 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 3.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |