Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
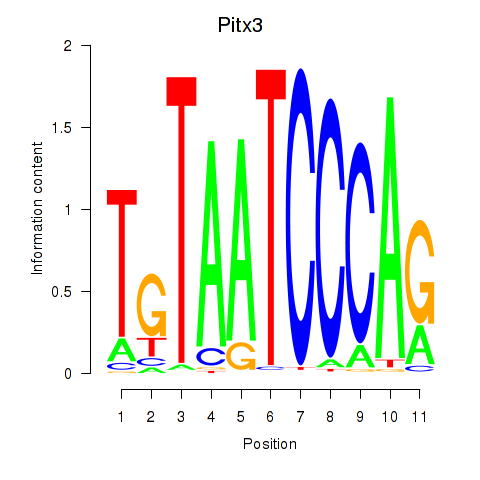
Results for Pitx3
Z-value: 1.72
Transcription factors associated with Pitx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx3
|
ENSMUSG00000025229.9 | paired-like homeodomain transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx3 | mm10_v2_chr19_-_46148369_46148386 | 0.36 | 3.3e-02 | Click! |
Activity profile of Pitx3 motif
Sorted Z-values of Pitx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_181051232 | 8.08 |
ENSMUST00000036819.6
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr9_+_119357381 | 7.02 |
ENSMUST00000039610.8
|
Xylb
|
xylulokinase homolog (H. influenzae) |
| chr8_-_72212837 | 6.67 |
ENSMUST00000098630.3
|
Cib3
|
calcium and integrin binding family member 3 |
| chr8_+_114133557 | 6.48 |
ENSMUST00000073521.5
ENSMUST00000066514.6 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr17_-_12675833 | 6.36 |
ENSMUST00000024596.8
|
Slc22a1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr8_+_114133601 | 6.27 |
ENSMUST00000109109.1
|
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr15_+_9335550 | 6.20 |
ENSMUST00000072403.6
|
Ugt3a2
|
UDP glycosyltransferases 3 family, polypeptide A2 |
| chr8_+_114133635 | 6.17 |
ENSMUST00000147605.1
ENSMUST00000134593.1 |
Nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr7_+_26835305 | 5.54 |
ENSMUST00000005685.8
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5 |
| chr10_+_88459569 | 5.52 |
ENSMUST00000020252.3
ENSMUST00000125612.1 |
Sycp3
|
synaptonemal complex protein 3 |
| chr13_-_12464925 | 5.47 |
ENSMUST00000124888.1
|
Lgals8
|
lectin, galactose binding, soluble 8 |
| chr12_-_104044431 | 5.22 |
ENSMUST00000043915.3
|
Serpina12
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr2_-_24048857 | 5.07 |
ENSMUST00000114497.1
|
Hnmt
|
histamine N-methyltransferase |
| chr8_+_127447669 | 4.97 |
ENSMUST00000159511.1
|
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr8_+_70083509 | 4.88 |
ENSMUST00000007738.9
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr7_-_48843663 | 4.88 |
ENSMUST00000167786.2
|
Csrp3
|
cysteine and glycine-rich protein 3 |
| chr7_-_100662414 | 4.72 |
ENSMUST00000079176.6
|
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr7_-_100662315 | 4.61 |
ENSMUST00000151123.1
ENSMUST00000107047.2 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr6_+_71493850 | 4.52 |
ENSMUST00000064637.4
ENSMUST00000114178.1 |
Rnf103
|
ring finger protein 103 |
| chrX_+_59999436 | 4.45 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chrX_+_142226765 | 4.38 |
ENSMUST00000112916.2
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr8_-_27228581 | 4.26 |
ENSMUST00000121838.1
|
Adrb3
|
adrenergic receptor, beta 3 |
| chr5_-_65428354 | 4.17 |
ENSMUST00000131263.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr6_-_85933379 | 4.17 |
ENSMUST00000162660.1
|
Nat8b
|
N-acetyltransferase 8B |
| chr11_-_100770926 | 4.10 |
ENSMUST00000139341.1
ENSMUST00000017891.7 |
Ghdc
|
GH3 domain containing |
| chr7_-_19692596 | 4.03 |
ENSMUST00000108451.2
ENSMUST00000045035.4 |
Apoc1
|
apolipoprotein C-I |
| chr10_-_41709297 | 3.89 |
ENSMUST00000019955.9
ENSMUST00000099932.3 |
Ccdc162
|
coiled-coil domain containing 162 |
| chr14_+_66635251 | 3.77 |
ENSMUST00000159365.1
ENSMUST00000054661.1 ENSMUST00000159068.1 |
Adra1a
|
adrenergic receptor, alpha 1a |
| chr7_+_119900099 | 3.74 |
ENSMUST00000106516.1
|
Lyrm1
|
LYR motif containing 1 |
| chr17_-_46763576 | 3.66 |
ENSMUST00000041012.7
|
Ptcra
|
pre T cell antigen receptor alpha |
| chr10_+_62920630 | 3.64 |
ENSMUST00000044977.3
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr7_-_68275098 | 3.55 |
ENSMUST00000135564.1
|
Gm16157
|
predicted gene 16157 |
| chr9_-_54068346 | 3.32 |
ENSMUST00000098760.3
|
Tnfaip8l3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr8_+_46010596 | 3.31 |
ENSMUST00000110381.2
|
Lrp2bp
|
Lrp2 binding protein |
| chrX_+_140381971 | 3.29 |
ENSMUST00000044702.6
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr7_+_27029074 | 3.20 |
ENSMUST00000075552.5
|
Cyp2a12
|
cytochrome P450, family 2, subfamily a, polypeptide 12 |
| chr7_-_12998172 | 3.14 |
ENSMUST00000120903.1
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr9_-_15306116 | 3.01 |
ENSMUST00000178977.1
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene |
| chr9_+_46269069 | 2.93 |
ENSMUST00000034584.3
|
Apoa5
|
apolipoprotein A-V |
| chr7_+_131410601 | 2.91 |
ENSMUST00000015829.7
ENSMUST00000117518.1 |
Acadsb
|
acyl-Coenzyme A dehydrogenase, short/branched chain |
| chr7_-_12998140 | 2.89 |
ENSMUST00000032539.7
|
Slc27a5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr9_-_103305049 | 2.89 |
ENSMUST00000142540.1
|
1300017J02Rik
|
RIKEN cDNA 1300017J02 gene |
| chr7_+_26307190 | 2.87 |
ENSMUST00000098657.3
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4 |
| chr6_+_90550789 | 2.81 |
ENSMUST00000130418.1
ENSMUST00000032175.8 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr8_-_3467617 | 2.78 |
ENSMUST00000111081.3
ENSMUST00000118194.1 ENSMUST00000004686.6 |
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chrX_+_7909542 | 2.76 |
ENSMUST00000086274.2
|
Gm10490
|
predicted gene 10490 |
| chr7_+_102225812 | 2.73 |
ENSMUST00000142873.1
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr19_+_7494033 | 2.72 |
ENSMUST00000170373.1
|
Atl3
|
atlastin GTPase 3 |
| chr12_+_36090379 | 2.72 |
ENSMUST00000071825.5
|
Gm5434
|
predicted gene 5434 |
| chr11_+_9048575 | 2.69 |
ENSMUST00000043285.4
|
Gm11992
|
predicted gene 11992 |
| chr8_-_72435043 | 2.66 |
ENSMUST00000109974.1
|
Calr3
|
calreticulin 3 |
| chr14_-_25927672 | 2.64 |
ENSMUST00000185006.1
|
Tmem254a
|
transmembrane protein 254a |
| chr1_+_88070765 | 2.52 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr11_-_120727226 | 2.42 |
ENSMUST00000106148.3
ENSMUST00000026144.4 |
Dcxr
|
dicarbonyl L-xylulose reductase |
| chr11_+_102041509 | 2.41 |
ENSMUST00000123895.1
ENSMUST00000017453.5 ENSMUST00000107163.2 ENSMUST00000107164.2 |
Cd300lg
|
CD300 antigen like family member G |
| chr1_+_167308649 | 2.39 |
ENSMUST00000097473.4
|
Tmco1
|
transmembrane and coiled-coil domains 1 |
| chr5_+_120513102 | 2.38 |
ENSMUST00000111889.1
|
Slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr6_-_72617000 | 2.36 |
ENSMUST00000070524.4
|
Tgoln1
|
trans-golgi network protein |
| chr18_+_51117754 | 2.33 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr10_+_62920648 | 2.31 |
ENSMUST00000144459.1
|
Slc25a16
|
solute carrier family 25 (mitochondrial carrier, Graves disease autoantigen), member 16 |
| chr9_+_77941274 | 2.30 |
ENSMUST00000134072.1
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr4_-_109202217 | 2.28 |
ENSMUST00000160774.1
ENSMUST00000030288.7 ENSMUST00000162787.2 |
Osbpl9
|
oxysterol binding protein-like 9 |
| chr4_+_155582476 | 2.28 |
ENSMUST00000105612.1
|
Nadk
|
NAD kinase |
| chr6_-_128526703 | 2.26 |
ENSMUST00000143664.1
ENSMUST00000112132.1 ENSMUST00000032510.7 |
Pzp
|
pregnancy zone protein |
| chr1_-_184883218 | 2.25 |
ENSMUST00000048308.5
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr16_-_91618986 | 2.24 |
ENSMUST00000143058.1
ENSMUST00000049244.8 ENSMUST00000169982.1 ENSMUST00000133731.1 |
Dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr9_+_114731177 | 2.22 |
ENSMUST00000035007.8
|
Cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr6_+_149582012 | 2.16 |
ENSMUST00000144085.2
|
Gm21814
|
predicted gene, 21814 |
| chr3_+_95318782 | 2.15 |
ENSMUST00000139866.1
|
Cers2
|
ceramide synthase 2 |
| chr7_-_126922887 | 2.13 |
ENSMUST00000134134.1
ENSMUST00000119781.1 ENSMUST00000121612.2 |
Tmem219
|
transmembrane protein 219 |
| chr15_+_99392882 | 2.12 |
ENSMUST00000023749.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr9_-_73039697 | 2.11 |
ENSMUST00000184035.1
ENSMUST00000098566.4 |
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr1_-_175979114 | 2.11 |
ENSMUST00000104983.1
|
B020018G12Rik
|
RIKEN cDNA B020018G12 gene |
| chr11_-_79296906 | 2.11 |
ENSMUST00000068448.2
|
Gm9964
|
predicted gene 9964 |
| chr2_-_32704123 | 2.10 |
ENSMUST00000127812.1
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr11_+_67025144 | 2.10 |
ENSMUST00000079077.5
ENSMUST00000061786.5 |
Tmem220
|
transmembrane protein 220 |
| chr10_+_58497918 | 2.07 |
ENSMUST00000036576.8
|
Ccdc138
|
coiled-coil domain containing 138 |
| chr4_-_19922599 | 2.07 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr14_-_26207041 | 2.05 |
ENSMUST00000022418.8
|
Tmem254b
|
transmembrane protein 254b |
| chr14_+_26082245 | 2.05 |
ENSMUST00000168149.2
ENSMUST00000163305.1 |
Cphx2
Cphx3
|
cytoplasmic polyadenylated homeobox 2 cytoplasmic polyadenylated homeobox 3 |
| chr9_-_73039644 | 2.02 |
ENSMUST00000184389.1
|
Pigb
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chrX_-_162888426 | 1.99 |
ENSMUST00000033723.3
|
Syap1
|
synapse associated protein 1 |
| chr9_-_64341145 | 1.96 |
ENSMUST00000120760.1
ENSMUST00000168844.2 |
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr7_+_68275970 | 1.94 |
ENSMUST00000153805.1
|
Fam169b
|
family with sequence similarity 169, member B |
| chr14_+_26221856 | 1.90 |
ENSMUST00000184577.1
|
Cphx3
|
cytoplasmic polyadenylated homeobox 3 |
| chr14_+_25942492 | 1.90 |
ENSMUST00000184016.1
|
Cphx1
|
cytoplasmic polyadenylated homeobox 1 |
| chr11_-_106580594 | 1.88 |
ENSMUST00000153870.1
|
Tex2
|
testis expressed gene 2 |
| chr1_+_128244122 | 1.88 |
ENSMUST00000027592.3
|
Ubxn4
|
UBX domain protein 4 |
| chr2_+_163122605 | 1.87 |
ENSMUST00000144092.1
|
Gm11454
|
predicted gene 11454 |
| chr19_+_43782181 | 1.85 |
ENSMUST00000026208.4
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr7_+_25619404 | 1.84 |
ENSMUST00000077338.5
ENSMUST00000085953.3 |
Atp5sl
|
ATP5S-like |
| chr6_+_56017489 | 1.84 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr17_-_32189457 | 1.82 |
ENSMUST00000087721.3
ENSMUST00000162117.1 |
Ephx3
|
epoxide hydrolase 3 |
| chr2_-_132578244 | 1.82 |
ENSMUST00000110142.1
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr5_+_92376954 | 1.82 |
ENSMUST00000120781.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr10_-_68278713 | 1.81 |
ENSMUST00000020106.7
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr4_+_141368116 | 1.79 |
ENSMUST00000006380.4
|
Fam131c
|
family with sequence similarity 131, member C |
| chr9_-_64341288 | 1.77 |
ENSMUST00000068367.7
|
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr13_-_74482943 | 1.74 |
ENSMUST00000074369.6
|
Zfp825
|
zinc finger protein 825 |
| chr9_-_20815048 | 1.73 |
ENSMUST00000004201.7
|
Col5a3
|
collagen, type V, alpha 3 |
| chr10_-_20548361 | 1.71 |
ENSMUST00000164195.1
|
Pde7b
|
phosphodiesterase 7B |
| chr7_-_7299492 | 1.71 |
ENSMUST00000000619.6
|
Clcn4-2
|
chloride channel 4-2 |
| chr10_+_80702649 | 1.71 |
ENSMUST00000095426.3
|
Izumo4
|
IZUMO family member 4 |
| chr10_+_128499364 | 1.69 |
ENSMUST00000180477.1
|
A430046D13Rik
|
Riken cDNA A430046D13 gene |
| chr1_-_59161594 | 1.66 |
ENSMUST00000078874.7
ENSMUST00000066374.7 |
Mpp4
|
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr1_-_134361933 | 1.65 |
ENSMUST00000049470.4
|
Tmem183a
|
transmembrane protein 183A |
| chr13_+_67779754 | 1.64 |
ENSMUST00000164936.2
ENSMUST00000181319.1 |
Zfp493
|
zinc finger protein 493 |
| chr19_-_37330613 | 1.64 |
ENSMUST00000131070.1
|
Ide
|
insulin degrading enzyme |
| chr2_+_144594054 | 1.63 |
ENSMUST00000136628.1
|
Gm561
|
predicted gene 561 |
| chr10_-_12861735 | 1.63 |
ENSMUST00000076817.4
|
Utrn
|
utrophin |
| chr10_-_20548320 | 1.63 |
ENSMUST00000169404.1
|
Pde7b
|
phosphodiesterase 7B |
| chr17_-_32420965 | 1.62 |
ENSMUST00000170392.1
|
Pglyrp2
|
peptidoglycan recognition protein 2 |
| chr5_+_30853796 | 1.61 |
ENSMUST00000126284.1
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chrX_-_166479633 | 1.60 |
ENSMUST00000049435.8
|
Rab9
|
RAB9, member RAS oncogene family |
| chr4_+_41755210 | 1.60 |
ENSMUST00000108038.1
ENSMUST00000084695.4 |
Galt
|
galactose-1-phosphate uridyl transferase |
| chr8_+_105708270 | 1.60 |
ENSMUST00000013302.5
|
4933405L10Rik
|
RIKEN cDNA 4933405L10 gene |
| chr11_-_120617887 | 1.59 |
ENSMUST00000106188.3
ENSMUST00000026129.9 |
Pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr10_-_77089428 | 1.59 |
ENSMUST00000156009.1
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr7_+_97720839 | 1.58 |
ENSMUST00000070021.1
|
Gm9990
|
predicted gene 9990 |
| chr6_-_85762480 | 1.57 |
ENSMUST00000168531.1
|
Cml3
|
camello-like 3 |
| chr8_-_105758570 | 1.56 |
ENSMUST00000155038.2
ENSMUST00000013294.9 |
Gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr4_-_133529326 | 1.56 |
ENSMUST00000163919.1
|
Gm17688
|
predicted gene, 17688 |
| chr1_+_185332143 | 1.55 |
ENSMUST00000027916.6
ENSMUST00000151769.1 ENSMUST00000110965.1 |
Bpnt1
|
bisphosphate 3'-nucleotidase 1 |
| chr8_+_36489191 | 1.55 |
ENSMUST00000171777.1
|
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr15_+_31572179 | 1.55 |
ENSMUST00000161088.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr3_-_88455735 | 1.54 |
ENSMUST00000029700.5
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr9_+_90054529 | 1.54 |
ENSMUST00000143172.1
|
Ctsh
|
cathepsin H |
| chrX_-_36874111 | 1.54 |
ENSMUST00000047486.5
|
C330007P06Rik
|
RIKEN cDNA C330007P06 gene |
| chr11_-_40695203 | 1.53 |
ENSMUST00000101347.3
|
Mat2b
|
methionine adenosyltransferase II, beta |
| chr4_-_44066960 | 1.51 |
ENSMUST00000173234.1
ENSMUST00000173274.1 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr18_-_3309858 | 1.51 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr11_-_98775333 | 1.51 |
ENSMUST00000064941.6
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr11_-_121388186 | 1.50 |
ENSMUST00000106107.2
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr2_-_132578155 | 1.50 |
ENSMUST00000110136.1
ENSMUST00000124107.1 ENSMUST00000060955.5 |
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr1_-_39651165 | 1.50 |
ENSMUST00000053355.4
|
Creg2
|
cellular repressor of E1A-stimulated genes 2 |
| chr17_-_35121990 | 1.47 |
ENSMUST00000173915.1
ENSMUST00000172765.2 |
Csnk2b
|
casein kinase 2, beta polypeptide |
| chr11_-_60811228 | 1.47 |
ENSMUST00000018744.8
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr11_-_4160286 | 1.46 |
ENSMUST00000093381.4
ENSMUST00000101626.2 |
Ccdc157
|
coiled-coil domain containing 157 |
| chr3_-_153912966 | 1.45 |
ENSMUST00000089950.4
|
Rabggtb
|
RAB geranylgeranyl transferase, b subunit |
| chr11_-_73324616 | 1.44 |
ENSMUST00000021119.2
|
Aspa
|
aspartoacylase |
| chr11_-_116131073 | 1.44 |
ENSMUST00000106440.2
ENSMUST00000067632.3 |
Trim65
|
tripartite motif-containing 65 |
| chr18_-_3309723 | 1.44 |
ENSMUST00000136961.1
ENSMUST00000152108.1 |
Crem
|
cAMP responsive element modulator |
| chr1_-_36244245 | 1.43 |
ENSMUST00000046875.7
|
Uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr10_+_80151154 | 1.43 |
ENSMUST00000146516.1
ENSMUST00000144526.1 |
Midn
|
midnolin |
| chr8_+_70131327 | 1.43 |
ENSMUST00000095273.5
|
Nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr5_+_107830244 | 1.43 |
ENSMUST00000072578.6
|
Ube2d2b
|
ubiquitin-conjugating enzyme E2D 2B |
| chr11_-_96916448 | 1.41 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr7_+_30169861 | 1.41 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr5_-_134456702 | 1.41 |
ENSMUST00000073161.5
ENSMUST00000171794.2 ENSMUST00000111245.2 ENSMUST00000100654.3 ENSMUST00000167084.2 ENSMUST00000100652.3 ENSMUST00000100650.3 ENSMUST00000074114.5 |
Gtf2ird1
|
general transcription factor II I repeat domain-containing 1 |
| chrY_+_818646 | 1.40 |
ENSMUST00000115894.1
|
Uba1y
|
ubiquitin-activating enzyme, Chr Y |
| chr5_+_106609098 | 1.40 |
ENSMUST00000167618.1
|
Gm17304
|
predicted gene, 17304 |
| chr7_-_141475131 | 1.40 |
ENSMUST00000043870.8
|
Polr2l
|
polymerase (RNA) II (DNA directed) polypeptide L |
| chr7_-_118584669 | 1.39 |
ENSMUST00000044195.4
|
Tmc7
|
transmembrane channel-like gene family 7 |
| chr4_-_12087912 | 1.38 |
ENSMUST00000050686.3
|
Tmem67
|
transmembrane protein 67 |
| chr9_-_108567336 | 1.37 |
ENSMUST00000074208.4
|
Ndufaf3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3 |
| chr19_-_29648355 | 1.35 |
ENSMUST00000159692.1
|
Ermp1
|
endoplasmic reticulum metallopeptidase 1 |
| chr14_-_20702991 | 1.34 |
ENSMUST00000180987.1
|
6230400D17Rik
|
RIKEN cDNA 6230400D17 gene |
| chr1_-_59237093 | 1.34 |
ENSMUST00000163058.1
ENSMUST00000027178.6 |
Als2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr6_-_83656082 | 1.33 |
ENSMUST00000014686.2
|
Clec4f
|
C-type lectin domain family 4, member f |
| chrX_+_36874351 | 1.32 |
ENSMUST00000016452.7
|
Ube2a
|
ubiquitin-conjugating enzyme E2A |
| chr13_-_41220395 | 1.32 |
ENSMUST00000021793.7
|
Elovl2
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
| chr1_-_90967667 | 1.32 |
ENSMUST00000131428.1
|
Rab17
|
RAB17, member RAS oncogene family |
| chr14_+_79397772 | 1.31 |
ENSMUST00000022600.2
|
Mtrf1
|
mitochondrial translational release factor 1 |
| chr8_-_71511762 | 1.31 |
ENSMUST00000048452.4
|
Plvap
|
plasmalemma vesicle associated protein |
| chr7_+_24257644 | 1.30 |
ENSMUST00000072713.6
|
Zfp108
|
zinc finger protein 108 |
| chr4_-_142015056 | 1.30 |
ENSMUST00000105780.1
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_+_51261719 | 1.30 |
ENSMUST00000130641.1
|
Clk4
|
CDC like kinase 4 |
| chr8_+_33653238 | 1.29 |
ENSMUST00000033992.8
|
Gsr
|
glutathione reductase |
| chr11_-_78080360 | 1.28 |
ENSMUST00000021183.3
|
Eral1
|
Era (G-protein)-like 1 (E. coli) |
| chr7_+_6252710 | 1.27 |
ENSMUST00000121024.1
|
Gm6792
|
predicted gene 6792 |
| chr7_+_19489045 | 1.27 |
ENSMUST00000011407.7
ENSMUST00000137613.1 |
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr2_+_69670100 | 1.27 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr5_-_121618865 | 1.27 |
ENSMUST00000041252.6
ENSMUST00000111776.1 |
Acad12
|
acyl-Coenzyme A dehydrogenase family, member 12 |
| chr5_+_117976761 | 1.27 |
ENSMUST00000035579.6
|
Fbxo21
|
F-box protein 21 |
| chr10_-_43540945 | 1.26 |
ENSMUST00000147196.1
ENSMUST00000019932.3 |
1700021F05Rik
|
RIKEN cDNA 1700021F05 gene |
| chr2_+_140152043 | 1.26 |
ENSMUST00000104994.2
|
Gm17374
|
predicted gene, 17374 |
| chr10_+_39612934 | 1.26 |
ENSMUST00000019987.6
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr11_+_22971991 | 1.26 |
ENSMUST00000049506.5
|
Zrsr1
|
zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1 |
| chr8_+_71951038 | 1.25 |
ENSMUST00000131237.1
ENSMUST00000136516.1 ENSMUST00000109997.2 ENSMUST00000132848.1 |
Zfp961
|
zinc finger protein 961 |
| chr17_+_46711459 | 1.25 |
ENSMUST00000002840.8
|
Pex6
|
peroxisomal biogenesis factor 6 |
| chr9_-_62070606 | 1.24 |
ENSMUST00000034785.7
|
Glce
|
glucuronyl C5-epimerase |
| chr3_-_88455692 | 1.24 |
ENSMUST00000107531.1
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr2_-_25236787 | 1.24 |
ENSMUST00000091318.4
|
Gm757
|
predicted gene 757 |
| chr11_-_29247208 | 1.23 |
ENSMUST00000020754.3
|
Ccdc104
|
coiled-coil domain containing 104 |
| chr8_-_72213636 | 1.23 |
ENSMUST00000109987.1
|
Gm11034
|
predicted gene 11034 |
| chr2_-_121380940 | 1.22 |
ENSMUST00000038389.8
|
Strc
|
stereocilin |
| chr11_+_101448403 | 1.21 |
ENSMUST00000010502.6
|
Ifi35
|
interferon-induced protein 35 |
| chr15_+_76701455 | 1.21 |
ENSMUST00000019224.7
|
Mfsd3
|
major facilitator superfamily domain containing 3 |
| chr10_+_61417972 | 1.20 |
ENSMUST00000049339.6
|
Nodal
|
nodal |
| chr8_+_47824459 | 1.20 |
ENSMUST00000038693.6
|
Cldn22
|
claudin 22 |
| chr9_+_22454290 | 1.19 |
ENSMUST00000168332.1
|
Gm17545
|
predicted gene, 17545 |
| chr6_+_114643106 | 1.19 |
ENSMUST00000169310.3
ENSMUST00000182169.1 ENSMUST00000183165.1 ENSMUST00000182098.1 ENSMUST00000182793.1 ENSMUST00000182902.1 ENSMUST00000182428.1 ENSMUST00000182035.1 |
Atg7
|
autophagy related 7 |
| chr7_-_144738520 | 1.19 |
ENSMUST00000118556.2
ENSMUST00000033393.8 |
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr2_-_132578128 | 1.19 |
ENSMUST00000028822.7
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr2_+_158502633 | 1.19 |
ENSMUST00000109484.1
|
Adig
|
adipogenin |
| chr1_+_171225054 | 1.18 |
ENSMUST00000111321.1
ENSMUST00000005824.5 ENSMUST00000111320.1 ENSMUST00000111319.1 |
Apoa2
|
apolipoprotein A-II |
| chr11_+_97029925 | 1.18 |
ENSMUST00000021249.4
|
Scrn2
|
secernin 2 |
| chr1_-_5070281 | 1.17 |
ENSMUST00000147158.1
ENSMUST00000118000.1 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr17_+_94873986 | 1.17 |
ENSMUST00000108007.4
|
Gm20939
|
predicted gene, 20939 |
| chr3_-_88334428 | 1.17 |
ENSMUST00000107552.1
|
Tmem79
|
transmembrane protein 79 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.9 | GO:0034031 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 3.1 | 9.4 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 1.6 | 4.9 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 1.5 | 6.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 1.5 | 5.8 | GO:0010248 | establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 1.4 | 5.5 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 1.3 | 3.8 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 1.1 | 4.3 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 1.0 | 4.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.0 | 5.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.9 | 2.8 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.9 | 5.5 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.8 | 5.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.7 | 3.7 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.7 | 2.2 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.7 | 2.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.7 | 2.0 | GO:0042335 | cuticle development(GO:0042335) |
| 0.7 | 2.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.5 | 2.1 | GO:1903976 | peripheral nervous system axon regeneration(GO:0014012) negative regulation of glial cell migration(GO:1903976) |
| 0.5 | 1.6 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.5 | 1.0 | GO:0010899 | regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.5 | 0.5 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.5 | 14.4 | GO:0080184 | response to phenylpropanoid(GO:0080184) |
| 0.5 | 1.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 2.8 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.5 | 1.8 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.5 | 2.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.3 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.4 | 2.1 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 1.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.4 | 1.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 1.6 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.4 | 5.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.4 | 1.2 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.4 | 1.5 | GO:0010813 | neuropeptide catabolic process(GO:0010813) dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.4 | 1.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 1.5 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.4 | 1.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.4 | 1.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 1.4 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.3 | 1.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.3 | 1.6 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.3 | 1.6 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.3 | 4.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.3 | 3.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.3 | 3.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 1.5 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.3 | 1.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 1.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.3 | 0.8 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 1.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.3 | 4.1 | GO:0097502 | mannosylation(GO:0097502) |
| 0.3 | 0.8 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.3 | 0.8 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 2.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.2 | 1.2 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.2 | 1.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 0.7 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.2 | 1.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.2 | 0.7 | GO:0046340 | transcription initiation from RNA polymerase I promoter(GO:0006361) diacylglycerol catabolic process(GO:0046340) |
| 0.2 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 1.6 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 1.8 | GO:0060613 | fat pad development(GO:0060613) |
| 0.2 | 3.1 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.2 | 2.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 1.1 | GO:0060295 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.6 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.2 | 1.5 | GO:0000237 | leptotene(GO:0000237) |
| 0.2 | 2.5 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.2 | 0.6 | GO:1905223 | epicardium morphogenesis(GO:1905223) |
| 0.2 | 2.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.2 | 0.6 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 2.4 | GO:0006983 | ER overload response(GO:0006983) |
| 0.2 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.8 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 0.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 1.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 3.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.6 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.2 | 1.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 0.6 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.2 | 1.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 1.3 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 0.7 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.7 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.2 | 1.4 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 0.5 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.2 | 1.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 0.3 | GO:0071898 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 0.6 | GO:0051790 | threonine metabolic process(GO:0006566) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.8 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.2 | 2.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.4 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.2 | 0.6 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.6 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.7 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.1 | 0.6 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.6 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) negative regulation of Fas signaling pathway(GO:1902045) |
| 0.1 | 1.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.8 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 0.9 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.8 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 1.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.5 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.5 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 0.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 1.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.0 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 1.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 3.5 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 3.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.6 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.1 | 0.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 3.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.1 | 0.3 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.1 | 1.5 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.3 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.1 | 4.2 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.1 | 0.6 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 3.1 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.6 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 1.0 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 2.5 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 1.3 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.1 | 0.9 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 2.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.1 | 6.6 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 0.3 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.9 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.3 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.1 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.1 | 0.9 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 1.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.2 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 1.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.3 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 2.7 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.8 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.2 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 1.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.8 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.5 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.5 | GO:2000002 | negative regulation of macrophage differentiation(GO:0045650) negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.1 | 0.9 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 1.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.5 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 4.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 0.7 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 | 0.3 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.1 | 1.0 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 1.3 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.1 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 1.8 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.4 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 | 0.8 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.1 | 0.8 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.4 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 2.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 1.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.5 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.7 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 1.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.6 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 5.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.9 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.6 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 1.3 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.2 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.0 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.6 | GO:0035404 | histone-serine phosphorylation(GO:0035404) |
| 0.0 | 2.9 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 1.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 1.1 | GO:0097286 | iron ion import(GO:0097286) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 2.0 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 3.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.6 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 1.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 1.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.5 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0036015 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 1.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.8 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 2.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.8 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.6 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.0 | 0.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to diacyl bacterial lipopeptide(GO:0071726) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.7 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.7 | GO:0035418 | protein localization to synapse(GO:0035418) |
| 0.0 | 4.2 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 4.6 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.9 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) antigen processing and presentation of peptide antigen(GO:0048002) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.0 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.6 | 1.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 2.3 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.5 | 1.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.4 | 1.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.4 | 1.6 | GO:0000802 | transverse filament(GO:0000802) |
| 0.3 | 1.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.3 | 3.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.3 | 1.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 1.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.3 | 6.8 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.2 | 0.9 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 1.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.2 | 1.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 2.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 0.8 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 2.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 0.6 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.2 | 2.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.2 | 0.9 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 2.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 2.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.7 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 1.5 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.9 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 2.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 2.7 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.1 | 2.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 10.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 22.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 7.8 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 13.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.7 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 2.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 1.7 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.1 | 0.1 | GO:0071010 | prespliceosome(GO:0071010) |
| 0.1 | 2.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 9.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.8 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 5.1 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 11.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.6 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 4.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.0 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 18.9 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 2.1 | 6.4 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 2.0 | 8.1 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 1.4 | 4.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 1.1 | 4.5 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.1 | 4.3 | GO:0031699 | beta-adrenergic receptor activity(GO:0004939) beta-3 adrenergic receptor binding(GO:0031699) |
| 0.9 | 3.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.9 | 2.8 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.8 | 4.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.7 | 2.2 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.7 | 2.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.6 | 2.4 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.5 | 4.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.5 | 1.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.5 | 6.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 1.4 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.4 | 1.6 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.4 | 1.5 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.4 | 2.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 2.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.4 | 1.4 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 2.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.4 | 1.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 2.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 1.6 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.3 | 11.6 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.3 | 1.5 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.3 | 3.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 1.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 1.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.3 | 0.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.3 | 1.6 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 2.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 1.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.3 | 1.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 2.3 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.3 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 2.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.7 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 0.7 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.2 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 8.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 1.8 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 2.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 4.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.1 | GO:0070287 | ferritin receptor activity(GO:0070287) |
| 0.2 | 1.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.8 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.8 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.2 | 1.3 | GO:0005534 | galactose binding(GO:0005534) |
| 0.2 | 4.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 1.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 4.1 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 1.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 1.3 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 2.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 0.6 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.7 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.1 | 1.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 1.4 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.6 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.4 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) U6 snRNA 3'-end binding(GO:0030629) |
| 0.1 | 1.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 1.8 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 4.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.0 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.4 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.1 | 4.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.9 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.6 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 4.4 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 2.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 1.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 1.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 3.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.5 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.6 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 0.3 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.1 | 1.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 4.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 1.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.6 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 1.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 1.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 5.4 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 2.4 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.1 | 1.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 0.3 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 1.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.3 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 1.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.4 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 4.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.6 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.6 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.8 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.1 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.9 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 5.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 7.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.9 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 2.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 1.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.1 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 2.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 4.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 2.6 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 2.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 5.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 3.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.6 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 4.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.8 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 5.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 3.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.5 | 6.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.5 | 6.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.4 | 5.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.3 | 4.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 8.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 3.6 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 4.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.2 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 2.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 5.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.0 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 0.9 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.9 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.6 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 3.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.6 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.7 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |