Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
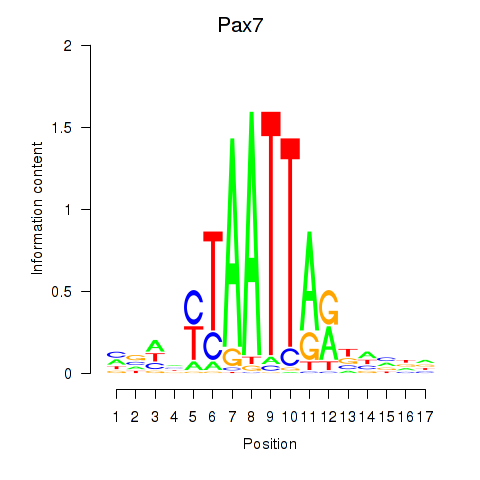
Results for Pax7
Z-value: 1.14
Transcription factors associated with Pax7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax7
|
ENSMUSG00000028736.7 | paired box 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax7 | mm10_v2_chr4_-_139833524_139833535 | 0.43 | 8.9e-03 | Click! |
Activity profile of Pax7 motif
Sorted Z-values of Pax7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_15924928 | 5.43 |
ENSMUST00000025542.3
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr19_-_15924560 | 5.38 |
ENSMUST00000162053.1
|
Psat1
|
phosphoserine aminotransferase 1 |
| chr5_-_138170992 | 5.28 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr10_-_62379852 | 3.94 |
ENSMUST00000143236.1
ENSMUST00000133429.1 ENSMUST00000132926.1 ENSMUST00000116238.2 |
Hk1
|
hexokinase 1 |
| chr12_+_117843489 | 3.77 |
ENSMUST00000021592.9
|
Cdca7l
|
cell division cycle associated 7 like |
| chr9_-_20959785 | 3.30 |
ENSMUST00000177754.1
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chrX_+_9885622 | 2.98 |
ENSMUST00000067529.2
ENSMUST00000086165.3 |
Sytl5
|
synaptotagmin-like 5 |
| chr6_+_123262107 | 2.85 |
ENSMUST00000032240.2
|
Clec4d
|
C-type lectin domain family 4, member d |
| chr1_-_52952834 | 2.56 |
ENSMUST00000050567.4
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr9_+_119063429 | 2.36 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr11_-_87359011 | 2.24 |
ENSMUST00000055438.4
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing) |
| chr15_+_9436028 | 2.21 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr13_+_76579670 | 2.18 |
ENSMUST00000126960.1
ENSMUST00000109583.2 |
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr11_-_102946688 | 2.05 |
ENSMUST00000057849.5
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr4_-_99654983 | 1.60 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr19_+_8802486 | 1.47 |
ENSMUST00000172175.1
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr17_-_35516780 | 1.44 |
ENSMUST00000160885.1
ENSMUST00000159009.1 ENSMUST00000161012.1 |
Tcf19
|
transcription factor 19 |
| chr3_-_75270073 | 1.44 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr11_-_99244058 | 1.43 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr18_-_77047243 | 1.42 |
ENSMUST00000137354.1
ENSMUST00000137498.1 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr3_-_17230976 | 1.38 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr18_-_77047282 | 1.38 |
ENSMUST00000154665.1
ENSMUST00000026486.6 ENSMUST00000123650.1 ENSMUST00000126153.1 |
Katnal2
|
katanin p60 subunit A-like 2 |
| chr13_-_22042949 | 1.34 |
ENSMUST00000091741.4
|
Hist1h2ag
|
histone cluster 1, H2ag |
| chr19_-_55241236 | 1.29 |
ENSMUST00000069183.6
|
Gucy2g
|
guanylate cyclase 2g |
| chr13_+_22043189 | 1.27 |
ENSMUST00000110452.1
|
Hist1h2bj
|
histone cluster 1, H2bj |
| chr13_+_23533869 | 1.22 |
ENSMUST00000073261.2
|
Hist1h2af
|
histone cluster 1, H2af |
| chr2_+_78051155 | 1.12 |
ENSMUST00000145972.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr2_+_105126505 | 1.12 |
ENSMUST00000143043.1
|
Wt1
|
Wilms tumor 1 homolog |
| chr6_-_87533219 | 1.11 |
ENSMUST00000113637.2
ENSMUST00000071024.6 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr3_-_87174657 | 1.05 |
ENSMUST00000159976.1
ENSMUST00000107618.2 |
Kirrel
|
kin of IRRE like (Drosophila) |
| chr2_+_125068118 | 1.01 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr13_+_55445301 | 0.99 |
ENSMUST00000001115.8
ENSMUST00000099482.3 |
Grk6
|
G protein-coupled receptor kinase 6 |
| chr2_+_86007778 | 0.95 |
ENSMUST00000062166.1
|
Olfr1032
|
olfactory receptor 1032 |
| chr11_+_43682038 | 0.95 |
ENSMUST00000094294.4
|
Pwwp2a
|
PWWP domain containing 2A |
| chr13_+_76579681 | 0.94 |
ENSMUST00000109589.2
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr13_-_102906046 | 0.81 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_26817357 | 0.74 |
ENSMUST00000071500.6
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr4_+_136286061 | 0.69 |
ENSMUST00000069195.4
ENSMUST00000130658.1 |
Zfp46
|
zinc finger protein 46 |
| chr9_+_38877126 | 0.68 |
ENSMUST00000078289.2
|
Olfr926
|
olfactory receptor 926 |
| chr11_+_43681998 | 0.67 |
ENSMUST00000061070.5
|
Pwwp2a
|
PWWP domain containing 2A |
| chr6_+_6248659 | 0.66 |
ENSMUST00000181633.1
ENSMUST00000176283.1 ENSMUST00000175814.1 ENSMUST00000181192.1 |
Gm20619
|
predicted gene 20619 |
| chr14_+_53324632 | 0.65 |
ENSMUST00000178100.1
|
Trav7n-6
|
T cell receptor alpha variable 7N-6 |
| chr7_+_45621805 | 0.65 |
ENSMUST00000033100.4
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr8_-_34965631 | 0.64 |
ENSMUST00000033929.4
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr2_-_85675173 | 0.58 |
ENSMUST00000099917.1
|
Olfr1006
|
olfactory receptor 1006 |
| chr14_+_32785963 | 0.58 |
ENSMUST00000039191.6
|
1810011H11Rik
|
RIKEN cDNA 1810011H11 gene |
| chr16_+_58408443 | 0.57 |
ENSMUST00000046663.7
|
Dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr6_+_92816460 | 0.57 |
ENSMUST00000057977.3
|
A730049H05Rik
|
RIKEN cDNA A730049H05 gene |
| chr19_-_11283813 | 0.55 |
ENSMUST00000067673.6
|
Ms4a5
|
membrane-spanning 4-domains, subfamily A, member 5 |
| chr2_+_85979312 | 0.54 |
ENSMUST00000170610.1
|
Olfr1030
|
olfactory receptor 1030 |
| chr7_-_19715395 | 0.51 |
ENSMUST00000032555.9
ENSMUST00000093552.5 |
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr14_-_69707546 | 0.47 |
ENSMUST00000118374.1
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr1_-_55226768 | 0.47 |
ENSMUST00000027121.8
ENSMUST00000114428.2 |
Rftn2
|
raftlin family member 2 |
| chr2_+_32727682 | 0.43 |
ENSMUST00000113242.2
|
Sh2d3c
|
SH2 domain containing 3C |
| chr1_+_72284367 | 0.42 |
ENSMUST00000027380.5
ENSMUST00000141783.1 |
Tmem169
|
transmembrane protein 169 |
| chr13_+_75707484 | 0.41 |
ENSMUST00000001583.6
|
Ell2
|
elongation factor RNA polymerase II 2 |
| chr11_+_99857915 | 0.40 |
ENSMUST00000107434.1
|
Gm11568
|
predicted gene 11568 |
| chr14_-_69707493 | 0.38 |
ENSMUST00000121142.1
|
R3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr9_-_96719404 | 0.37 |
ENSMUST00000140121.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr2_+_78051220 | 0.37 |
ENSMUST00000144728.1
|
4930440I19Rik
|
RIKEN cDNA 4930440I19 gene |
| chr3_-_106790143 | 0.36 |
ENSMUST00000038845.8
|
Cd53
|
CD53 antigen |
| chr2_+_86041317 | 0.36 |
ENSMUST00000111589.1
|
Olfr1033
|
olfactory receptor 1033 |
| chr10_+_85386813 | 0.35 |
ENSMUST00000105307.1
ENSMUST00000020231.3 |
Btbd11
|
BTB (POZ) domain containing 11 |
| chr13_-_102905740 | 0.33 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_96268654 | 0.28 |
ENSMUST00000098843.2
|
Hist2h3b
|
histone cluster 2, H3b |
| chr8_+_107031218 | 0.27 |
ENSMUST00000034388.9
|
Vps4a
|
vacuolar protein sorting 4a (yeast) |
| chr7_-_46667375 | 0.27 |
ENSMUST00000107669.2
|
Tph1
|
tryptophan hydroxylase 1 |
| chr3_-_72967854 | 0.26 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr2_-_116067391 | 0.24 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr7_+_27486910 | 0.24 |
ENSMUST00000008528.7
|
Sertad1
|
SERTA domain containing 1 |
| chr12_+_72441852 | 0.19 |
ENSMUST00000162159.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr13_-_113042243 | 0.16 |
ENSMUST00000099162.2
|
Gm10735
|
predicted gene 10735 |
| chr2_-_144270852 | 0.15 |
ENSMUST00000110030.3
|
Snx5
|
sorting nexin 5 |
| chr10_-_24092320 | 0.13 |
ENSMUST00000092654.2
|
Taar8b
|
trace amine-associated receptor 8B |
| chr12_+_72441933 | 0.13 |
ENSMUST00000161284.1
|
Lrrc9
|
leucine rich repeat containing 9 |
| chr9_-_96719549 | 0.12 |
ENSMUST00000128269.1
|
Zbtb38
|
zinc finger and BTB domain containing 38 |
| chr18_+_77332394 | 0.07 |
ENSMUST00000148341.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr2_+_86046451 | 0.02 |
ENSMUST00000079298.2
|
Olfr1034
|
olfactory receptor 1034 |
| chr4_-_14621805 | 0.01 |
ENSMUST00000042221.7
|
Slc26a7
|
solute carrier family 26, member 7 |
| chr17_+_35517100 | 0.00 |
ENSMUST00000164242.2
ENSMUST00000045956.7 |
Cchcr1
|
coiled-coil alpha-helical rod protein 1 |
| chr2_+_27165233 | 0.00 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr2_-_150255591 | 0.00 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 10.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.8 | 3.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.6 | 2.8 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.5 | 5.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.5 | 2.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.5 | 3.9 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.4 | 1.1 | GO:2001074 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.3 | 1.0 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 | 1.0 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 0.6 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 2.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:1903774 | mitotic cytokinesis checkpoint(GO:0044878) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 3.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.4 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.5 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.1 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 1.4 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 2.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.6 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 2.9 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 1.0 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.7 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 0.7 | GO:1901381 | positive regulation of potassium ion transmembrane transport(GO:1901381) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.4 | 5.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 3.9 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 3.3 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 3.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 3.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 10.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.4 | 2.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 1.0 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 1.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 5.3 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 2.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 2.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 3.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 3.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.0 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 3.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 3.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 4.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 5.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 3.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |