Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
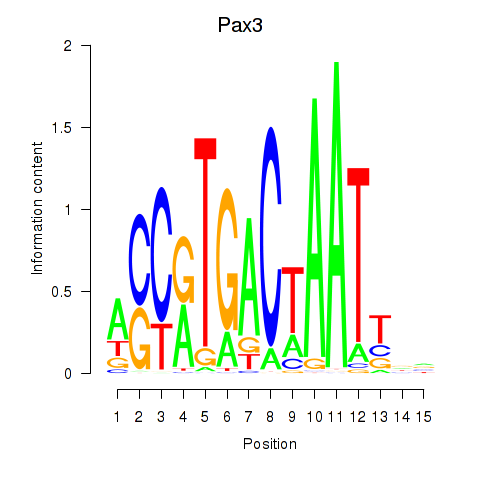
Results for Pax3
Z-value: 1.06
Transcription factors associated with Pax3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax3
|
ENSMUSG00000004872.9 | paired box 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax3 | mm10_v2_chr1_-_78196832_78196845 | 0.28 | 9.4e-02 | Click! |
Activity profile of Pax3 motif
Sorted Z-values of Pax3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_84167466 | 6.38 |
ENSMUST00000050771.7
|
Gm11437
|
predicted gene 11437 |
| chr19_-_46148369 | 4.67 |
ENSMUST00000026259.9
|
Pitx3
|
paired-like homeodomain transcription factor 3 |
| chr2_+_102659213 | 4.62 |
ENSMUST00000111213.1
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_-_87092546 | 4.46 |
ENSMUST00000132667.1
ENSMUST00000145617.1 ENSMUST00000094649.4 |
Ugt2b36
|
UDP glucuronosyltransferase 2 family, polypeptide B36 |
| chr13_+_4434306 | 3.81 |
ENSMUST00000021630.8
|
Akr1c6
|
aldo-keto reductase family 1, member C6 |
| chr7_-_133782721 | 1.61 |
ENSMUST00000063669.1
|
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr4_-_25281801 | 1.44 |
ENSMUST00000102994.3
|
Ufl1
|
UFM1 specific ligase 1 |
| chr14_+_51893610 | 1.43 |
ENSMUST00000047726.5
ENSMUST00000161888.1 |
Slc39a2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr11_-_88851462 | 1.42 |
ENSMUST00000107903.1
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr7_-_31111148 | 1.38 |
ENSMUST00000164929.1
|
Hpn
|
hepsin |
| chr19_-_36736653 | 1.32 |
ENSMUST00000087321.2
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C |
| chr4_-_25281752 | 1.03 |
ENSMUST00000038705.7
|
Ufl1
|
UFM1 specific ligase 1 |
| chr4_+_127077374 | 0.94 |
ENSMUST00000046751.6
ENSMUST00000094713.3 |
Zmym6
|
zinc finger, MYM-type 6 |
| chr8_-_71043040 | 0.87 |
ENSMUST00000170101.1
|
Gm17576
|
predicted gene, 17576 |
| chr17_+_21691860 | 0.78 |
ENSMUST00000072133.4
|
Gm10226
|
predicted gene 10226 |
| chr1_-_130887408 | 0.75 |
ENSMUST00000121040.1
|
Il24
|
interleukin 24 |
| chr10_+_82138253 | 0.73 |
ENSMUST00000105314.2
|
AU041133
|
expressed sequence AU041133 |
| chr10_-_128923948 | 0.73 |
ENSMUST00000131271.1
|
Bloc1s1
|
biogenesis of lysosome-related organelles complex-1, subunit 1 |
| chr8_+_33428709 | 0.72 |
ENSMUST00000059351.7
|
5930422O12Rik
|
RIKEN cDNA 5930422O12 gene |
| chr2_+_133552159 | 0.66 |
ENSMUST00000028836.6
|
Bmp2
|
bone morphogenetic protein 2 |
| chr6_-_48766519 | 0.66 |
ENSMUST00000038811.8
|
Gimap3
|
GTPase, IMAP family member 3 |
| chr13_-_21716143 | 0.65 |
ENSMUST00000091756.1
|
Hist1h2bl
|
histone cluster 1, H2bl |
| chr3_+_36159522 | 0.63 |
ENSMUST00000165956.2
|
D3Ertd254e
|
DNA segment, Chr 3, ERATO Doi 254, expressed |
| chr12_-_72184990 | 0.58 |
ENSMUST00000021494.4
|
Ccdc175
|
coiled-coil domain containing 175 |
| chr8_+_110618577 | 0.57 |
ENSMUST00000034190.9
|
Vac14
|
Vac14 homolog (S. cerevisiae) |
| chr16_+_20548577 | 0.55 |
ENSMUST00000003319.5
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr16_-_32877723 | 0.54 |
ENSMUST00000119810.1
|
1700021K19Rik
|
RIKEN cDNA 1700021K19 gene |
| chr13_+_14630237 | 0.52 |
ENSMUST00000178289.1
ENSMUST00000038690.4 |
AW209491
|
expressed sequence AW209491 |
| chr10_-_128923439 | 0.52 |
ENSMUST00000153731.1
ENSMUST00000026405.3 |
Bloc1s1
|
biogenesis of lysosome-related organelles complex-1, subunit 1 |
| chr16_+_32877775 | 0.49 |
ENSMUST00000023489.4
ENSMUST00000171325.1 |
Fyttd1
|
forty-two-three domain containing 1 |
| chr1_+_176814660 | 0.49 |
ENSMUST00000056773.8
ENSMUST00000027785.8 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr4_-_63745055 | 0.47 |
ENSMUST00000062246.6
|
Tnfsf15
|
tumor necrosis factor (ligand) superfamily, member 15 |
| chr11_-_59571813 | 0.46 |
ENSMUST00000071943.2
|
Olfr222
|
olfactory receptor 222 |
| chr11_+_70970467 | 0.45 |
ENSMUST00000178822.1
ENSMUST00000108529.3 ENSMUST00000169965.1 ENSMUST00000167509.1 |
Rpain
|
RPA interacting protein |
| chr13_+_24327415 | 0.42 |
ENSMUST00000167746.1
|
Cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr9_+_64235201 | 0.39 |
ENSMUST00000039011.3
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr6_+_34920971 | 0.37 |
ENSMUST00000185102.1
ENSMUST00000114997.2 |
Stra8
|
stimulated by retinoic acid gene 8 |
| chr3_+_108186332 | 0.34 |
ENSMUST00000050909.6
ENSMUST00000106659.2 ENSMUST00000106656.1 ENSMUST00000106661.2 |
Amigo1
|
adhesion molecule with Ig like domain 1 |
| chr18_+_35562158 | 0.34 |
ENSMUST00000166793.1
|
Matr3
|
matrin 3 |
| chr3_-_75451818 | 0.31 |
ENSMUST00000178270.1
|
Wdr49
|
WD repeat domain 49 |
| chr16_-_88056176 | 0.27 |
ENSMUST00000072256.5
ENSMUST00000023652.8 ENSMUST00000114137.1 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr8_+_71887264 | 0.23 |
ENSMUST00000034259.7
|
Zfp709
|
zinc finger protein 709 |
| chr10_+_104194042 | 0.18 |
ENSMUST00000181615.1
|
Gm4340
|
predicted gene 4340 |
| chr10_+_104142987 | 0.18 |
ENSMUST00000181634.1
|
Gm6763
|
predicted gene 6763 |
| chr10_+_104151497 | 0.18 |
ENSMUST00000181287.1
|
Gm6763
|
predicted gene 6763 |
| chr10_+_104160006 | 0.18 |
ENSMUST00000181179.1
|
Gm8764
|
predicted gene 8764 |
| chr10_+_104168515 | 0.18 |
ENSMUST00000181059.1
|
Gm21304
|
predicted gene, 21304 |
| chr10_+_104177024 | 0.18 |
ENSMUST00000180889.1
|
Gm21312
|
predicted gene, 21312 |
| chr10_+_104185533 | 0.18 |
ENSMUST00000181703.1
|
Gm20765
|
predicted gene, 20765 |
| chr18_+_34759551 | 0.16 |
ENSMUST00000097622.3
|
Fam53c
|
family with sequence similarity 53, member C |
| chr10_+_104194457 | 0.16 |
ENSMUST00000181036.1
|
Gm4340
|
predicted gene 4340 |
| chr10_+_104143402 | 0.16 |
ENSMUST00000180664.1
|
Gm6763
|
predicted gene 6763 |
| chr10_+_104151912 | 0.16 |
ENSMUST00000180692.1
|
Gm6763
|
predicted gene 6763 |
| chr10_+_104160421 | 0.16 |
ENSMUST00000181166.1
|
Gm8764
|
predicted gene 8764 |
| chr10_+_104168930 | 0.16 |
ENSMUST00000180568.1
|
Gm21304
|
predicted gene, 21304 |
| chr10_+_104177439 | 0.16 |
ENSMUST00000181239.1
|
Gm21312
|
predicted gene, 21312 |
| chr10_+_104185948 | 0.16 |
ENSMUST00000181707.1
|
Gm20765
|
predicted gene, 20765 |
| chr3_-_59262825 | 0.15 |
ENSMUST00000050360.7
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr18_+_34758890 | 0.14 |
ENSMUST00000049281.5
|
Fam53c
|
family with sequence similarity 53, member C |
| chr17_+_37529957 | 0.14 |
ENSMUST00000097325.3
|
Olfr111
|
olfactory receptor 111 |
| chr19_+_34008247 | 0.13 |
ENSMUST00000054260.6
|
Lipk
|
lipase, family member K |
| chr17_+_87107621 | 0.09 |
ENSMUST00000041369.6
|
Socs5
|
suppressor of cytokine signaling 5 |
| chrX_+_114474312 | 0.09 |
ENSMUST00000113371.1
ENSMUST00000040504.5 |
Klhl4
|
kelch-like 4 |
| chrX_-_103623704 | 0.05 |
ENSMUST00000130063.1
ENSMUST00000125419.1 |
Ftx
|
Ftx transcript, Xist regulator (non-protein coding) |
| chr10_-_107912134 | 0.05 |
ENSMUST00000165341.3
|
Otogl
|
otogelin-like |
| chr2_-_136387929 | 0.03 |
ENSMUST00000035264.2
ENSMUST00000077200.3 |
Pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 1.3 | 3.8 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.7 | 4.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.4 | 2.5 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 1.4 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 1.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 0.7 | GO:0035054 | BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) embryonic heart tube anterior/posterior pattern specification(GO:0035054) corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.1 | 0.4 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 | 0.3 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 | 1.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.4 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 1.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.7 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 1.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.3 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 4.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.6 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.8 | 2.5 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.8 | 4.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0030338 | CMP-N-acetylneuraminate monooxygenase activity(GO:0030338) |
| 0.1 | 4.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.7 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.6 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |