Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr6a1
Z-value: 1.10
Transcription factors associated with Nr6a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr6a1
|
ENSMUSG00000063972.7 | nuclear receptor subfamily 6, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr6a1 | mm10_v2_chr2_-_38926217_38926454 | 0.13 | 4.4e-01 | Click! |
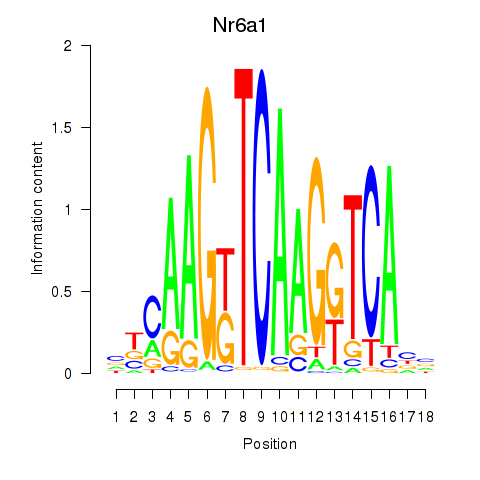
Activity profile of Nr6a1 motif
Sorted Z-values of Nr6a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_107230608 | 4.46 |
ENSMUST00000179399.1
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr16_+_22951072 | 3.26 |
ENSMUST00000023590.8
|
Hrg
|
histidine-rich glycoprotein |
| chr17_-_57228003 | 3.00 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr6_+_42245907 | 2.54 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr7_-_105600103 | 2.39 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr9_+_73101836 | 2.12 |
ENSMUST00000172578.1
|
Khdc3
|
KH domain containing 3, subcortical maternal complex member |
| chr1_+_160978576 | 2.06 |
ENSMUST00000064725.5
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr6_-_124636085 | 1.85 |
ENSMUST00000068797.2
|
Gm5077
|
predicted gene 5077 |
| chr9_-_119157055 | 1.85 |
ENSMUST00000010795.4
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr5_+_92376954 | 1.66 |
ENSMUST00000120781.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr6_+_90333270 | 1.42 |
ENSMUST00000164761.1
ENSMUST00000046128.9 |
Uroc1
|
urocanase domain containing 1 |
| chr15_+_31572179 | 1.42 |
ENSMUST00000161088.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr4_-_82505274 | 1.40 |
ENSMUST00000050872.8
ENSMUST00000064770.2 |
Nfib
|
nuclear factor I/B |
| chrX_-_8018492 | 1.37 |
ENSMUST00000033503.2
|
Glod5
|
glyoxalase domain containing 5 |
| chr2_+_25054396 | 1.36 |
ENSMUST00000102931.4
ENSMUST00000074422.7 ENSMUST00000132172.1 ENSMUST00000114388.1 ENSMUST00000114386.1 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr9_+_122805524 | 1.32 |
ENSMUST00000052740.7
|
Tcaim
|
T cell activation inhibitor, mitochondrial |
| chr9_+_53301571 | 1.32 |
ENSMUST00000051014.1
|
Exph5
|
exophilin 5 |
| chr4_-_82505707 | 1.28 |
ENSMUST00000107248.1
ENSMUST00000107247.1 |
Nfib
|
nuclear factor I/B |
| chr14_+_78849171 | 1.27 |
ENSMUST00000040990.5
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr11_-_60811228 | 1.25 |
ENSMUST00000018744.8
|
Shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr7_-_140881811 | 1.24 |
ENSMUST00000106048.3
ENSMUST00000147331.2 ENSMUST00000137710.1 |
Sirt3
|
sirtuin 3 |
| chr12_-_28623282 | 1.18 |
ENSMUST00000036136.7
|
Colec11
|
collectin sub-family member 11 |
| chr1_+_131970589 | 1.11 |
ENSMUST00000027695.6
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr2_+_127425155 | 1.08 |
ENSMUST00000062211.3
ENSMUST00000110373.1 |
Gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr10_+_21978643 | 1.06 |
ENSMUST00000142174.1
ENSMUST00000164659.1 |
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr5_-_108674753 | 1.04 |
ENSMUST00000119270.1
ENSMUST00000163328.1 ENSMUST00000136227.1 |
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr13_+_25056004 | 0.97 |
ENSMUST00000036932.8
|
Dcdc2a
|
doublecortin domain containing 2a |
| chr4_+_138434647 | 0.95 |
ENSMUST00000044058.4
ENSMUST00000105813.1 ENSMUST00000105815.1 |
Mul1
|
mitochondrial ubiquitin ligase activator of NFKB 1 |
| chr10_-_68541842 | 0.93 |
ENSMUST00000020103.2
|
1700040L02Rik
|
RIKEN cDNA 1700040L02 gene |
| chr14_+_12189943 | 0.91 |
ENSMUST00000119888.1
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr11_-_100770926 | 0.91 |
ENSMUST00000139341.1
ENSMUST00000017891.7 |
Ghdc
|
GH3 domain containing |
| chr4_-_82505749 | 0.88 |
ENSMUST00000107245.2
ENSMUST00000107246.1 |
Nfib
|
nuclear factor I/B |
| chr4_-_45530330 | 0.87 |
ENSMUST00000061986.5
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr4_+_155562348 | 0.84 |
ENSMUST00000030939.7
|
Nadk
|
NAD kinase |
| chr9_-_50603792 | 0.83 |
ENSMUST00000000175.4
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr2_+_32606946 | 0.79 |
ENSMUST00000113290.1
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr2_+_32606979 | 0.75 |
ENSMUST00000113289.1
ENSMUST00000095044.3 |
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
| chr6_-_57535422 | 0.68 |
ENSMUST00000042766.3
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing) |
| chr2_+_180071793 | 0.63 |
ENSMUST00000108901.1
|
Mtg2
|
mitochondrial ribosome associated GTPase 2 |
| chr17_-_46645128 | 0.62 |
ENSMUST00000003642.6
|
Klc4
|
kinesin light chain 4 |
| chr17_-_34804546 | 0.59 |
ENSMUST00000025223.8
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr6_-_113434529 | 0.59 |
ENSMUST00000133348.1
|
Cidec
|
cell death-inducing DFFA-like effector c |
| chr18_+_64887690 | 0.54 |
ENSMUST00000163516.1
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr11_-_99244058 | 0.53 |
ENSMUST00000103132.3
ENSMUST00000038214.6 |
Krt222
|
keratin 222 |
| chr5_-_137611429 | 0.52 |
ENSMUST00000031731.7
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr5_-_137610626 | 0.52 |
ENSMUST00000142675.1
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr6_+_48593927 | 0.47 |
ENSMUST00000135151.1
|
Repin1
|
replication initiator 1 |
| chr9_+_73102398 | 0.46 |
ENSMUST00000034737.6
ENSMUST00000173734.2 ENSMUST00000167514.1 ENSMUST00000174203.2 |
Khdc3
Gm20509
|
KH domain containing 3, subcortical maternal complex member predicted gene 20509 |
| chr2_+_177768044 | 0.45 |
ENSMUST00000108942.3
|
Gm14322
|
predicted gene 14322 |
| chr5_-_137611372 | 0.42 |
ENSMUST00000054564.6
|
Pcolce
|
procollagen C-endopeptidase enhancer protein |
| chr15_-_58364148 | 0.39 |
ENSMUST00000068515.7
|
Anxa13
|
annexin A13 |
| chr8_-_109693235 | 0.39 |
ENSMUST00000034164.4
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr12_-_115790884 | 0.39 |
ENSMUST00000081809.5
|
Ighv1-73
|
immunoglobulin heavy variable 1-73 |
| chr19_+_46707443 | 0.37 |
ENSMUST00000003655.7
|
As3mt
|
arsenic (+3 oxidation state) methyltransferase |
| chr4_-_123904826 | 0.36 |
ENSMUST00000181292.1
|
Gm26606
|
predicted gene, 26606 |
| chr11_-_76763550 | 0.35 |
ENSMUST00000010536.8
|
Gosr1
|
golgi SNAP receptor complex member 1 |
| chrX_+_140381971 | 0.34 |
ENSMUST00000044702.6
|
Frmpd3
|
FERM and PDZ domain containing 3 |
| chr6_-_129622685 | 0.34 |
ENSMUST00000032252.5
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr4_+_43632185 | 0.32 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr17_+_72918298 | 0.32 |
ENSMUST00000024857.6
|
Lbh
|
limb-bud and heart |
| chr10_-_43540945 | 0.26 |
ENSMUST00000147196.1
ENSMUST00000019932.3 |
1700021F05Rik
|
RIKEN cDNA 1700021F05 gene |
| chr11_+_120677226 | 0.26 |
ENSMUST00000129644.2
ENSMUST00000151160.1 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr12_-_71136611 | 0.24 |
ENSMUST00000021486.8
ENSMUST00000166120.1 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr9_-_121839460 | 0.24 |
ENSMUST00000135986.2
|
Ccdc13
|
coiled-coil domain containing 13 |
| chr13_+_113794505 | 0.20 |
ENSMUST00000091201.6
|
Arl15
|
ADP-ribosylation factor-like 15 |
| chr10_+_23970661 | 0.20 |
ENSMUST00000092659.2
|
Taar5
|
trace amine-associated receptor 5 |
| chr2_+_172472512 | 0.20 |
ENSMUST00000029007.2
|
Fam209
|
family with sequence similarity 209 |
| chr2_+_176431829 | 0.19 |
ENSMUST00000172025.1
|
Gm14435
|
predicted gene 14435 |
| chr1_+_151428612 | 0.18 |
ENSMUST00000065625.5
|
Trmt1l
|
tRNA methyltransferase 1 like |
| chr9_+_87144285 | 0.17 |
ENSMUST00000113149.1
ENSMUST00000143779.1 ENSMUST00000179313.1 ENSMUST00000049457.7 ENSMUST00000153444.1 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr9_+_50603892 | 0.14 |
ENSMUST00000044051.4
|
Timm8b
|
translocase of inner mitochondrial membrane 8B |
| chr10_+_128083273 | 0.14 |
ENSMUST00000026459.5
|
Atp5b
|
ATP synthase, H+ transporting mitochondrial F1 complex, beta subunit |
| chr5_-_136986829 | 0.13 |
ENSMUST00000034953.7
ENSMUST00000085941.5 |
Znhit1
|
zinc finger, HIT domain containing 1 |
| chr15_-_98165613 | 0.12 |
ENSMUST00000143400.1
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr5_-_123572976 | 0.10 |
ENSMUST00000031388.8
|
Vps33a
|
vacuolar protein sorting 33A (yeast) |
| chr7_+_16944645 | 0.09 |
ENSMUST00000094807.5
|
Pnmal2
|
PNMA-like 2 |
| chr5_+_30232581 | 0.09 |
ENSMUST00000145167.1
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr3_-_96402010 | 0.06 |
ENSMUST00000181412.1
|
4930477E14Rik
|
RIKEN cDNA 4930477E14 gene |
| chr11_-_69822144 | 0.06 |
ENSMUST00000045771.6
|
Spem1
|
sperm maturation 1 |
| chr14_+_76488436 | 0.05 |
ENSMUST00000101618.2
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr5_+_136987019 | 0.05 |
ENSMUST00000004968.4
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr1_+_171329015 | 0.04 |
ENSMUST00000111300.1
|
Dedd
|
death effector domain-containing |
| chr15_-_98165560 | 0.04 |
ENSMUST00000123922.1
|
Asb8
|
ankyrin repeat and SOCS box-containing 8 |
| chr1_+_171329376 | 0.02 |
ENSMUST00000111299.1
ENSMUST00000064950.4 |
Dedd
|
death effector domain-containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr6a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.9 | 3.6 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.6 | 3.0 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.3 | 1.4 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.3 | 2.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.3 | 1.4 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.3 | 1.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.2 | 0.9 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.2 | 1.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 0.8 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 1.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.1 | 1.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.3 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 1.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.4 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 1.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 2.6 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.1 | 0.5 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.0 | 2.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.3 | GO:1904672 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.5 | GO:2001273 | regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.2 | 0.8 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 2.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 3.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 1.0 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 1.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 3.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 3.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 1.2 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 0.3 | 1.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.3 | 0.8 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 1.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 1.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 3.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 1.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 1.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.0 | 3.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.9 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 4.4 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) endopeptidase regulator activity(GO:0061135) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 1.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.1 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 4.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 2.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 3.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |