Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
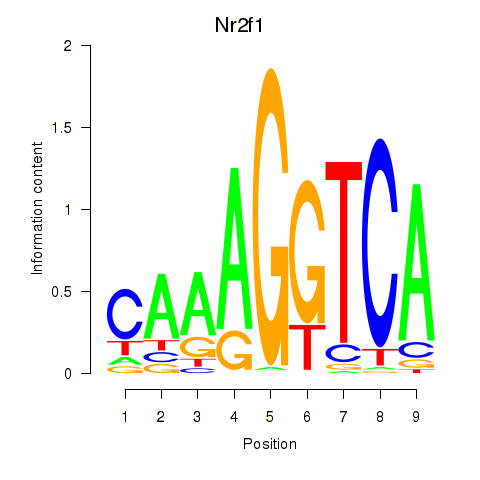
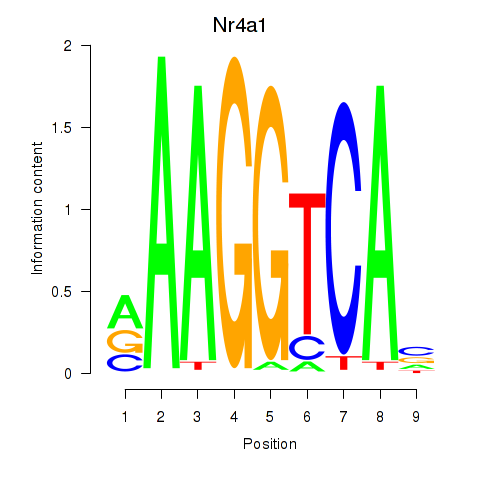
Results for Nr2f1_Nr4a1
Z-value: 2.37
Transcription factors associated with Nr2f1_Nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2f1
|
ENSMUSG00000069171.7 | nuclear receptor subfamily 2, group F, member 1 |
|
Nr4a1
|
ENSMUSG00000023034.6 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2f1 | mm10_v2_chr13_-_78196373_78196440 | -0.42 | 1.0e-02 | Click! |
| Nr4a1 | mm10_v2_chr15_+_101266839_101266859 | -0.18 | 3.0e-01 | Click! |
Activity profile of Nr2f1_Nr4a1 motif
Sorted Z-values of Nr2f1_Nr4a1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_46235260 | 30.12 |
ENSMUST00000121916.1
ENSMUST00000034586.2 |
Apoc3
|
apolipoprotein C-III |
| chr9_-_46235631 | 25.34 |
ENSMUST00000118649.1
|
Apoc3
|
apolipoprotein C-III |
| chr10_+_128194446 | 13.81 |
ENSMUST00000044776.6
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr7_-_141276729 | 11.07 |
ENSMUST00000167263.1
ENSMUST00000080654.5 |
Cdhr5
|
cadherin-related family member 5 |
| chr7_-_105600103 | 10.74 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr2_+_173153048 | 10.63 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr9_-_22002599 | 10.51 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr10_+_128194631 | 9.16 |
ENSMUST00000123291.1
|
Gls2
|
glutaminase 2 (liver, mitochondrial) |
| chr4_-_62087261 | 8.50 |
ENSMUST00000107488.3
ENSMUST00000107472.1 ENSMUST00000084531.4 |
Mup3
|
major urinary protein 3 |
| chr4_-_107307118 | 8.41 |
ENSMUST00000126291.1
ENSMUST00000106748.1 ENSMUST00000129138.1 ENSMUST00000082426.3 |
Dio1
|
deiodinase, iodothyronine, type I |
| chr11_+_115462464 | 8.39 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
| chr9_-_65422773 | 8.29 |
ENSMUST00000065894.5
|
Slc51b
|
solute carrier family 51, beta subunit |
| chr9_+_107340593 | 8.06 |
ENSMUST00000042581.2
|
6430571L13Rik
|
RIKEN cDNA 6430571L13 gene |
| chr2_+_71981184 | 7.83 |
ENSMUST00000090826.5
ENSMUST00000102698.3 |
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_166254095 | 7.59 |
ENSMUST00000111416.1
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr9_+_55326913 | 7.55 |
ENSMUST00000085754.3
ENSMUST00000034862.4 |
AI118078
|
expressed sequence AI118078 |
| chr6_+_90550789 | 7.40 |
ENSMUST00000130418.1
ENSMUST00000032175.8 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr19_-_4498574 | 7.27 |
ENSMUST00000048482.6
|
2010003K11Rik
|
RIKEN cDNA 2010003K11 gene |
| chr4_-_61303802 | 7.20 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr5_-_66151323 | 7.16 |
ENSMUST00000131838.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr11_+_75510077 | 6.73 |
ENSMUST00000042972.6
|
Rilp
|
Rab interacting lysosomal protein |
| chr9_-_21989427 | 6.73 |
ENSMUST00000045726.6
|
Rgl3
|
ral guanine nucleotide dissociation stimulator-like 3 |
| chr19_+_46131888 | 6.71 |
ENSMUST00000043739.3
|
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3 |
| chr17_-_32917320 | 6.60 |
ENSMUST00000179434.1
|
Cyp4f14
|
cytochrome P450, family 4, subfamily f, polypeptide 14 |
| chr9_-_70141484 | 6.57 |
ENSMUST00000034749.8
|
Fam81a
|
family with sequence similarity 81, member A |
| chr4_-_60662358 | 6.57 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr19_-_43524462 | 6.51 |
ENSMUST00000026196.7
|
Got1
|
glutamate oxaloacetate transaminase 1, soluble |
| chr1_+_88166004 | 6.48 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr4_-_141623799 | 6.44 |
ENSMUST00000038661.7
|
Slc25a34
|
solute carrier family 25, member 34 |
| chr19_-_39463067 | 6.26 |
ENSMUST00000035488.2
|
Cyp2c38
|
cytochrome P450, family 2, subfamily c, polypeptide 38 |
| chr11_-_113709520 | 6.18 |
ENSMUST00000173655.1
ENSMUST00000100248.4 |
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr19_+_37697792 | 6.16 |
ENSMUST00000025946.5
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr10_-_42478280 | 6.14 |
ENSMUST00000151747.1
|
Lace1
|
lactation elevated 1 |
| chr1_-_139781236 | 6.11 |
ENSMUST00000027612.8
ENSMUST00000111989.2 ENSMUST00000111986.2 |
Gm4788
|
predicted gene 4788 |
| chr6_-_119544282 | 6.02 |
ENSMUST00000119369.1
ENSMUST00000178696.1 |
Wnt5b
|
wingless-related MMTV integration site 5B |
| chr11_-_116198701 | 5.77 |
ENSMUST00000072948.4
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr4_-_61674094 | 5.63 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_60421933 | 5.62 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_+_134396320 | 5.60 |
ENSMUST00000105869.2
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr7_+_46751832 | 5.58 |
ENSMUST00000075982.2
|
Saa2
|
serum amyloid A 2 |
| chr4_-_60139857 | 5.55 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr4_-_62054112 | 5.52 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr4_+_140961203 | 5.50 |
ENSMUST00000010007.8
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr11_-_113708952 | 5.50 |
ENSMUST00000106617.1
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr1_+_167618246 | 5.49 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr18_+_45268876 | 5.46 |
ENSMUST00000183850.1
ENSMUST00000066890.7 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr4_-_60582152 | 5.46 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr3_-_138131356 | 5.45 |
ENSMUST00000029805.8
|
Mttp
|
microsomal triglyceride transfer protein |
| chr5_+_114146525 | 5.42 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr7_+_119617804 | 5.41 |
ENSMUST00000135683.1
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr19_-_8405060 | 5.41 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr18_-_35627223 | 5.40 |
ENSMUST00000025212.5
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1 |
| chr5_-_123182704 | 5.36 |
ENSMUST00000154713.1
ENSMUST00000031398.7 |
Hpd
|
4-hydroxyphenylpyruvic acid dioxygenase |
| chr10_-_42478488 | 5.28 |
ENSMUST00000041024.8
|
Lace1
|
lactation elevated 1 |
| chr4_-_61303998 | 5.25 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60222580 | 5.02 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr3_-_20275659 | 5.01 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr2_+_58755177 | 5.00 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr7_-_31055594 | 5.00 |
ENSMUST00000039909.6
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr3_+_32736990 | 4.94 |
ENSMUST00000127477.1
ENSMUST00000121778.1 ENSMUST00000154257.1 |
Ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 |
| chr17_-_56117577 | 4.93 |
ENSMUST00000019808.5
|
Plin5
|
perilipin 5 |
| chr17_-_56117265 | 4.90 |
ENSMUST00000113072.2
|
Plin5
|
perilipin 5 |
| chr3_+_130617645 | 4.89 |
ENSMUST00000163620.1
|
Etnppl
|
ethanolamine phosphate phospholyase |
| chr2_+_58754910 | 4.82 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chrX_-_38456407 | 4.67 |
ENSMUST00000074913.5
ENSMUST00000016678.7 ENSMUST00000061755.8 |
Lamp2
|
lysosomal-associated membrane protein 2 |
| chr9_+_46268601 | 4.64 |
ENSMUST00000121598.1
|
Apoa5
|
apolipoprotein A-V |
| chr1_+_74284930 | 4.59 |
ENSMUST00000113805.1
ENSMUST00000027370.6 ENSMUST00000087226.4 |
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia |
| chr1_-_140183283 | 4.53 |
ENSMUST00000111977.1
|
Cfh
|
complement component factor h |
| chr10_+_79716588 | 4.51 |
ENSMUST00000099513.1
ENSMUST00000020581.2 |
Hcn2
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 2 |
| chr11_-_116199040 | 4.48 |
ENSMUST00000066587.5
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl |
| chr2_-_25501717 | 4.47 |
ENSMUST00000015227.3
|
C8g
|
complement component 8, gamma polypeptide |
| chr1_-_140183404 | 4.44 |
ENSMUST00000066859.6
ENSMUST00000111976.2 |
Cfh
|
complement component factor h |
| chr5_-_146009598 | 4.40 |
ENSMUST00000138870.1
ENSMUST00000068317.6 |
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25 |
| chr13_-_74350206 | 4.39 |
ENSMUST00000022062.7
|
Sdha
|
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr9_+_108080436 | 4.37 |
ENSMUST00000035211.7
ENSMUST00000162886.1 |
Mst1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr11_-_72266596 | 4.36 |
ENSMUST00000021161.6
ENSMUST00000140167.1 |
Slc13a5
|
solute carrier family 13 (sodium-dependent citrate transporter), member 5 |
| chr1_-_139560158 | 4.35 |
ENSMUST00000160423.1
ENSMUST00000023965.5 |
Cfhr1
|
complement factor H-related 1 |
| chr17_+_43952999 | 4.27 |
ENSMUST00000177857.1
|
Rcan2
|
regulator of calcineurin 2 |
| chr4_+_133553370 | 4.27 |
ENSMUST00000042706.2
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr17_-_34804546 | 4.26 |
ENSMUST00000025223.8
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr4_-_6275629 | 4.24 |
ENSMUST00000029905.1
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1 |
| chr6_-_119467210 | 4.22 |
ENSMUST00000118120.1
|
Wnt5b
|
wingless-related MMTV integration site 5B |
| chr9_-_107668967 | 4.20 |
ENSMUST00000177567.1
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr6_+_42245907 | 4.18 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr10_-_78464969 | 4.18 |
ENSMUST00000041616.8
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr9_+_46240696 | 4.15 |
ENSMUST00000034585.6
|
Apoa4
|
apolipoprotein A-IV |
| chr6_-_113531575 | 4.06 |
ENSMUST00000032425.5
|
Emc3
|
ER membrane protein complex subunit 3 |
| chr4_-_61835185 | 4.02 |
ENSMUST00000082287.2
|
Mup5
|
major urinary protein 5 |
| chr5_-_24995748 | 4.01 |
ENSMUST00000076306.5
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr2_-_28563362 | 3.98 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr19_-_7966000 | 3.95 |
ENSMUST00000182102.1
ENSMUST00000075619.4 |
Slc22a27
|
solute carrier family 22, member 27 |
| chr17_-_73950172 | 3.94 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr2_-_91636394 | 3.80 |
ENSMUST00000111335.1
ENSMUST00000028681.8 |
F2
|
coagulation factor II |
| chr10_-_78464853 | 3.79 |
ENSMUST00000105385.1
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr3_-_98630309 | 3.77 |
ENSMUST00000044094.4
|
Hsd3b5
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 |
| chr7_-_31054815 | 3.77 |
ENSMUST00000071697.4
ENSMUST00000108110.3 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chrX_+_59999436 | 3.75 |
ENSMUST00000033477.4
|
F9
|
coagulation factor IX |
| chr4_-_148159571 | 3.73 |
ENSMUST00000167160.1
ENSMUST00000151246.1 |
Fbxo44
|
F-box protein 44 |
| chr12_+_108334341 | 3.71 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr6_+_91157373 | 3.70 |
ENSMUST00000155007.1
|
Hdac11
|
histone deacetylase 11 |
| chr8_+_94838321 | 3.66 |
ENSMUST00000034234.8
ENSMUST00000159871.1 |
Coq9
|
coenzyme Q9 homolog (yeast) |
| chr6_+_138140521 | 3.65 |
ENSMUST00000120939.1
ENSMUST00000120302.1 |
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr10_+_110920170 | 3.65 |
ENSMUST00000020403.5
|
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr8_+_70072896 | 3.61 |
ENSMUST00000110160.2
ENSMUST00000049197.5 |
Tm6sf2
|
transmembrane 6 superfamily member 2 |
| chrX_-_72656135 | 3.60 |
ENSMUST00000055966.6
|
Gabra3
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 3 |
| chr11_+_70054334 | 3.59 |
ENSMUST00000018699.6
ENSMUST00000108585.2 |
Asgr1
|
asialoglycoprotein receptor 1 |
| chr11_-_53423123 | 3.58 |
ENSMUST00000036045.5
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr11_-_53430779 | 3.58 |
ENSMUST00000061326.4
ENSMUST00000109021.3 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr14_+_75242287 | 3.57 |
ENSMUST00000022576.8
|
Cpb2
|
carboxypeptidase B2 (plasma) |
| chr1_-_139858684 | 3.56 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr4_-_129227883 | 3.55 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr4_+_117252010 | 3.53 |
ENSMUST00000125943.1
ENSMUST00000106434.1 |
Tmem53
|
transmembrane protein 53 |
| chr7_-_25477607 | 3.53 |
ENSMUST00000098669.1
ENSMUST00000098668.1 ENSMUST00000098666.2 |
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr4_+_60003438 | 3.52 |
ENSMUST00000107517.1
ENSMUST00000107520.1 |
Mup6
|
major urinary protein 6 |
| chr11_+_74649462 | 3.52 |
ENSMUST00000092915.5
ENSMUST00000117818.1 |
Cluh
|
clustered mitochondria (cluA/CLU1) homolog |
| chr8_-_72443772 | 3.51 |
ENSMUST00000019876.5
|
Calr3
|
calreticulin 3 |
| chr11_-_94677404 | 3.47 |
ENSMUST00000116349.2
|
Xylt2
|
xylosyltransferase II |
| chr10_-_115587739 | 3.47 |
ENSMUST00000020350.8
|
Lgr5
|
leucine rich repeat containing G protein coupled receptor 5 |
| chr1_+_131797381 | 3.46 |
ENSMUST00000112393.2
ENSMUST00000048660.5 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr10_-_20724696 | 3.44 |
ENSMUST00000170265.1
|
Pde7b
|
phosphodiesterase 7B |
| chr4_+_117251951 | 3.42 |
ENSMUST00000062824.5
|
Tmem53
|
transmembrane protein 53 |
| chr7_+_140920896 | 3.42 |
ENSMUST00000183845.1
ENSMUST00000106045.1 |
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr4_-_63662910 | 3.41 |
ENSMUST00000184252.1
|
Gm11214
|
predicted gene 11214 |
| chr11_-_11898044 | 3.39 |
ENSMUST00000066237.3
|
Ddc
|
dopa decarboxylase |
| chr7_+_140920940 | 3.37 |
ENSMUST00000184560.1
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr11_-_11898092 | 3.37 |
ENSMUST00000178704.1
|
Ddc
|
dopa decarboxylase |
| chr7_+_83631959 | 3.36 |
ENSMUST00000075418.7
ENSMUST00000117410.1 |
Stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr15_+_76343504 | 3.35 |
ENSMUST00000023210.6
|
Cyc1
|
cytochrome c-1 |
| chr2_-_166155624 | 3.34 |
ENSMUST00000109249.2
|
Sulf2
|
sulfatase 2 |
| chr18_-_39490649 | 3.33 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr7_-_127345314 | 3.31 |
ENSMUST00000060783.5
|
Zfp768
|
zinc finger protein 768 |
| chr4_+_138967112 | 3.30 |
ENSMUST00000116094.2
|
Rnf186
|
ring finger protein 186 |
| chr1_-_193264006 | 3.30 |
ENSMUST00000161737.1
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr8_+_36457548 | 3.29 |
ENSMUST00000135373.1
ENSMUST00000147525.1 |
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr10_+_4611971 | 3.29 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr1_-_130661584 | 3.25 |
ENSMUST00000137276.2
|
C4bp
|
complement component 4 binding protein |
| chr10_-_92375367 | 3.22 |
ENSMUST00000182870.1
|
Gm20757
|
predicted gene, 20757 |
| chr4_+_47288287 | 3.19 |
ENSMUST00000146967.1
|
Col15a1
|
collagen, type XV, alpha 1 |
| chr1_-_130661613 | 3.17 |
ENSMUST00000027657.7
|
C4bp
|
complement component 4 binding protein |
| chr13_-_41847482 | 3.16 |
ENSMUST00000072012.3
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr9_-_22085391 | 3.15 |
ENSMUST00000179422.1
ENSMUST00000098937.3 ENSMUST00000177967.1 ENSMUST00000180180.1 |
Ecsit
|
ECSIT homolog (Drosophila) |
| chr6_+_91156665 | 3.12 |
ENSMUST00000041736.4
|
Hdac11
|
histone deacetylase 11 |
| chr6_+_91156772 | 3.08 |
ENSMUST00000143621.1
|
Hdac11
|
histone deacetylase 11 |
| chr13_-_41847599 | 3.08 |
ENSMUST00000179758.1
|
Adtrp
|
androgen dependent TFPI regulating protein |
| chr10_-_20725023 | 3.07 |
ENSMUST00000020165.7
|
Pde7b
|
phosphodiesterase 7B |
| chr5_-_116422858 | 3.03 |
ENSMUST00000036991.4
|
Hspb8
|
heat shock protein 8 |
| chr2_-_25500613 | 3.02 |
ENSMUST00000040042.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr1_+_88138364 | 2.94 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr2_+_31887262 | 2.94 |
ENSMUST00000138325.1
ENSMUST00000028187.6 |
Lamc3
|
laminin gamma 3 |
| chr15_+_7129557 | 2.94 |
ENSMUST00000067190.5
ENSMUST00000164529.1 |
Lifr
|
leukemia inhibitory factor receptor |
| chr4_+_128993224 | 2.94 |
ENSMUST00000030583.6
ENSMUST00000102604.4 |
Ak2
|
adenylate kinase 2 |
| chr3_+_62338344 | 2.90 |
ENSMUST00000079300.6
|
Arhgef26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr16_-_20730544 | 2.86 |
ENSMUST00000076422.5
|
Thpo
|
thrombopoietin |
| chr4_-_59960659 | 2.86 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr19_-_46672883 | 2.86 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr17_-_34862473 | 2.84 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr6_+_41302265 | 2.83 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr8_+_119394866 | 2.83 |
ENSMUST00000098367.4
|
Mlycd
|
malonyl-CoA decarboxylase |
| chr3_+_118562129 | 2.81 |
ENSMUST00000039177.7
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr7_+_105554360 | 2.80 |
ENSMUST00000046983.8
|
Smpd1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr5_-_108674753 | 2.79 |
ENSMUST00000119270.1
ENSMUST00000163328.1 ENSMUST00000136227.1 |
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr10_+_39899304 | 2.76 |
ENSMUST00000181590.1
|
4930547M16Rik
|
RIKEN cDNA 4930547M16 gene |
| chr16_+_10545339 | 2.71 |
ENSMUST00000066345.7
ENSMUST00000115824.3 ENSMUST00000155633.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr16_-_23520579 | 2.71 |
ENSMUST00000089883.5
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr16_+_10545390 | 2.70 |
ENSMUST00000115827.1
ENSMUST00000038145.6 ENSMUST00000150894.1 |
Clec16a
|
C-type lectin domain family 16, member A |
| chr15_-_68363139 | 2.69 |
ENSMUST00000175699.1
|
Gm20732
|
predicted gene 20732 |
| chr4_+_43632185 | 2.68 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr12_+_41024329 | 2.67 |
ENSMUST00000134965.1
|
Immp2l
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr11_-_55033398 | 2.66 |
ENSMUST00000108883.3
ENSMUST00000102727.2 |
Anxa6
|
annexin A6 |
| chr17_-_34862122 | 2.65 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chr6_+_145211134 | 2.59 |
ENSMUST00000111725.1
ENSMUST00000111726.3 ENSMUST00000039729.3 ENSMUST00000111723.1 ENSMUST00000111724.1 ENSMUST00000111721.1 ENSMUST00000111719.1 |
Lyrm5
|
LYR motif containing 5 |
| chr7_-_79842287 | 2.56 |
ENSMUST00000049004.6
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr8_-_13494479 | 2.56 |
ENSMUST00000033828.5
|
Gas6
|
growth arrest specific 6 |
| chr7_-_81706905 | 2.55 |
ENSMUST00000026922.7
|
Homer2
|
homer homolog 2 (Drosophila) |
| chr8_+_45658666 | 2.54 |
ENSMUST00000134675.1
ENSMUST00000139869.1 ENSMUST00000126067.1 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr5_-_137921612 | 2.53 |
ENSMUST00000031741.7
|
Cyp3a13
|
cytochrome P450, family 3, subfamily a, polypeptide 13 |
| chr2_-_160872985 | 2.53 |
ENSMUST00000109460.1
ENSMUST00000127201.1 |
Zhx3
|
zinc fingers and homeoboxes 3 |
| chr5_-_66151903 | 2.50 |
ENSMUST00000167950.1
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_134554348 | 2.50 |
ENSMUST00000028704.2
|
Hao1
|
hydroxyacid oxidase 1, liver |
| chr7_+_44465391 | 2.50 |
ENSMUST00000035929.4
ENSMUST00000146128.1 |
Aspdh
|
aspartate dehydrogenase domain containing |
| chr5_-_147322435 | 2.48 |
ENSMUST00000100433.4
|
Urad
|
ureidoimidazoline (2-oxo-4-hydroxy-4-carboxy-5) decarboxylase |
| chr2_+_126556128 | 2.48 |
ENSMUST00000141482.2
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr16_+_13940630 | 2.48 |
ENSMUST00000141971.1
ENSMUST00000124947.1 ENSMUST00000023360.7 ENSMUST00000143697.1 |
Mpv17l
|
Mpv17 transgene, kidney disease mutant-like |
| chr13_+_64248649 | 2.48 |
ENSMUST00000181403.1
|
1810034E14Rik
|
RIKEN cDNA 1810034E14 gene |
| chr11_+_75468040 | 2.47 |
ENSMUST00000043598.7
ENSMUST00000108435.1 |
Tlcd2
|
TLC domain containing 2 |
| chr7_-_126897424 | 2.45 |
ENSMUST00000120007.1
|
Tmem219
|
transmembrane protein 219 |
| chr2_+_69670100 | 2.44 |
ENSMUST00000100050.3
|
Klhl41
|
kelch-like 41 |
| chr9_+_7445822 | 2.38 |
ENSMUST00000034497.6
|
Mmp3
|
matrix metallopeptidase 3 |
| chr15_+_54952939 | 2.37 |
ENSMUST00000181704.1
|
Gm26684
|
predicted gene, 26684 |
| chr17_-_25868727 | 2.36 |
ENSMUST00000026828.5
|
Fam195a
|
family with sequence similarity 195, member A |
| chr11_-_100704217 | 2.35 |
ENSMUST00000017974.6
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr1_-_134235420 | 2.35 |
ENSMUST00000038191.6
ENSMUST00000086465.4 |
Adora1
|
adenosine A1 receptor |
| chr4_+_134397380 | 2.34 |
ENSMUST00000105870.1
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr6_-_124542281 | 2.32 |
ENSMUST00000159463.1
ENSMUST00000162844.1 ENSMUST00000160505.1 ENSMUST00000162443.1 |
C1s
|
complement component 1, s subcomponent |
| chr6_-_59426279 | 2.32 |
ENSMUST00000051065.4
|
Gprin3
|
GPRIN family member 3 |
| chr14_-_51922773 | 2.31 |
ENSMUST00000089771.2
|
Rnase13
|
ribonuclease, RNase A family, 13 (non-active) |
| chr2_+_152105722 | 2.31 |
ENSMUST00000099225.2
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr1_-_136131171 | 2.30 |
ENSMUST00000146091.3
ENSMUST00000165464.1 ENSMUST00000166747.1 ENSMUST00000134998.1 |
Gm15850
|
predicted gene 15850 |
| chr11_+_96251100 | 2.29 |
ENSMUST00000129907.2
|
Gm53
|
predicted gene 53 |
| chr1_+_88200601 | 2.29 |
ENSMUST00000049289.8
|
Ugt1a2
|
UDP glucuronosyltransferase 1 family, polypeptide A2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2f1_Nr4a1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.9 | 55.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 4.9 | 14.8 | GO:0061402 | positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 3.3 | 20.0 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.7 | 8.2 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 2.7 | 8.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.7 | 8.0 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 2.5 | 7.4 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 2.5 | 9.8 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 2.2 | 11.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 2.1 | 10.7 | GO:0015886 | heme transport(GO:0015886) |
| 2.1 | 6.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 2.0 | 7.8 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 2.0 | 5.9 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 1.8 | 10.9 | GO:1903275 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 1.8 | 5.4 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 1.7 | 1.7 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 1.7 | 10.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 1.7 | 13.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.6 | 4.7 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 1.5 | 6.2 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 1.3 | 3.9 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 1.3 | 5.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.2 | 2.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 1.2 | 3.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 1.2 | 3.5 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 1.2 | 4.6 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 1.1 | 4.4 | GO:0015744 | succinate transport(GO:0015744) |
| 1.1 | 5.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 1.1 | 5.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.1 | 10.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 1.0 | 9.4 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 1.0 | 3.1 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 1.0 | 4.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 1.0 | 4.0 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 1.0 | 3.0 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.0 | 11.7 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 1.0 | 13.6 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 1.0 | 11.7 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 1.0 | 6.8 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 1.0 | 6.8 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.9 | 13.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.9 | 5.5 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.9 | 3.7 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.8 | 7.5 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.8 | 4.1 | GO:0060296 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.8 | 2.5 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.8 | 5.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.8 | 1.6 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.8 | 2.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.8 | 2.3 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.8 | 2.3 | GO:0032242 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.7 | 18.4 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.7 | 4.4 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.7 | 2.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.7 | 5.0 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.7 | 2.9 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.7 | 2.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.7 | 3.5 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.7 | 9.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.7 | 2.0 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.7 | 2.7 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.7 | 2.0 | GO:0001192 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.7 | 4.0 | GO:0051715 | cytolysis in other organism(GO:0051715) |
| 0.7 | 10.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.6 | 4.5 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.6 | 2.6 | GO:0097048 | positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.6 | 2.5 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.6 | 9.0 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.6 | 2.9 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.6 | 7.0 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.6 | 7.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.6 | 3.4 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.6 | 0.6 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.6 | 3.3 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.6 | 2.2 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.5 | 3.3 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.5 | 2.7 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 2.7 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.5 | 6.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.5 | 2.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.5 | 2.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.5 | 2.0 | GO:0046618 | drug export(GO:0046618) |
| 0.5 | 4.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.5 | 1.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.5 | 3.9 | GO:0006689 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.5 | 1.4 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.5 | 3.3 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.5 | 1.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.5 | 4.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 1.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.4 | 7.1 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.4 | 2.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.4 | 2.2 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.4 | 4.4 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.4 | 2.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.4 | 1.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.4 | 1.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.4 | 2.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.4 | 1.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 1.6 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.4 | 0.8 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.4 | 16.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.4 | 1.2 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.4 | 2.3 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.4 | 1.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.4 | 2.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.4 | 1.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.4 | 1.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 0.7 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.4 | 1.8 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.4 | 2.2 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.4 | 1.8 | GO:0009446 | spermidine biosynthetic process(GO:0008295) putrescine biosynthetic process(GO:0009446) |
| 0.4 | 2.8 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.4 | 1.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) high-density lipoprotein particle clearance(GO:0034384) |
| 0.4 | 2.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) termination of signal transduction(GO:0023021) |
| 0.3 | 2.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.3 | 1.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.3 | 3.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.3 | 1.0 | GO:0003360 | brainstem development(GO:0003360) |
| 0.3 | 1.0 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.3 | 2.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.3 | 1.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.3 | 1.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.3 | 1.9 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.3 | 1.6 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 | 0.3 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.3 | 11.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 3.3 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.3 | 3.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.3 | 1.8 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.3 | 1.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 | 2.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.3 | 1.7 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.3 | 0.9 | GO:1904925 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.3 | 1.4 | GO:0072363 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) |
| 0.3 | 1.7 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.3 | 0.8 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 2.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.3 | 2.7 | GO:1900194 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 1.6 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.3 | 1.3 | GO:0030300 | regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.3 | 1.8 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.3 | 0.8 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.3 | 0.8 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 1.5 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 6.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.2 | 2.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.2 | 1.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 3.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.2 | 1.9 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.2 | 0.7 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.2 | 1.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.2 | 2.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.9 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.2 | 0.7 | GO:0070973 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 0.7 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 1.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.2 | 2.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 3.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 2.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 1.7 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 2.6 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.2 | 1.0 | GO:0006547 | histidine metabolic process(GO:0006547) histidine catabolic process(GO:0006548) |
| 0.2 | 6.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.2 | 2.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.8 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.2 | 8.5 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.2 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 6.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.8 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.2 | 0.8 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.2 | 1.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 | 2.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.2 | 2.4 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.2 | 0.9 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.2 | 0.6 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 1.9 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.2 | 0.2 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.2 | 0.6 | GO:0051466 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of isotype switching to IgE isotypes(GO:0048295) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.2 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 0.2 | 2.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.9 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.2 | 0.9 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.2 | 6.6 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.2 | 2.7 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.2 | 1.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.2 | 3.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 | 0.7 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.2 | 1.0 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 0.7 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 1.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 2.7 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 | 2.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.7 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.2 | 1.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 1.0 | GO:0061525 | hindgut development(GO:0061525) |
| 0.2 | 1.0 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 | 1.9 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.2 | 3.8 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.2 | 1.7 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 5.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 1.8 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.2 | 0.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 1.0 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.6 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 2.4 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.1 | 0.4 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 4.7 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 3.4 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.1 | 0.7 | GO:0032344 | regulation of aldosterone metabolic process(GO:0032344) |
| 0.1 | 0.5 | GO:0043691 | reverse cholesterol transport(GO:0043691) |
| 0.1 | 0.8 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.8 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 1.4 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.1 | 2.0 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.1 | 0.3 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.1 | 0.8 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 2.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.1 | 2.7 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.5 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.2 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.1 | 0.7 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.1 | 1.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 3.7 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 3.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 1.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 2.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.6 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.1 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) |
| 0.1 | 0.3 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.1 | 0.5 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.5 | GO:0097490 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.4 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.4 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.7 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 1.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 2.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.2 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 1.0 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 11.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 6.8 | GO:0031016 | pancreas development(GO:0031016) |
| 0.1 | 0.8 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 1.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.8 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.1 | 1.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.5 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.1 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 0.1 | 0.9 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.7 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.6 | GO:1990000 | amyloid fibril formation(GO:1990000) |
| 0.1 | 0.8 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.1 | 5.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 4.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 | 1.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.4 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.1 | 2.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.1 | 0.6 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.1 | 9.1 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.1 | 1.1 | GO:0097237 | cellular response to toxic substance(GO:0097237) |
| 0.1 | 4.8 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.1 | 3.5 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.8 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.6 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 1.6 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.5 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 1.0 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 1.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 1.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 1.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.1 | GO:0046439 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.6 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 1.0 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 3.2 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.1 | 0.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.5 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 3.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.3 | GO:0071420 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.1 | 2.0 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.3 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.1 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.1 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.2 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.5 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.5 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 1.2 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.5 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.8 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.4 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.7 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.7 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 1.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 3.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 2.4 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 1.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 3.7 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.8 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 2.1 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) |
| 0.0 | 0.6 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.3 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.7 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.3 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 1.1 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.9 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.4 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 3.5 | GO:0006839 | mitochondrial transport(GO:0006839) |
| 0.0 | 0.3 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.0 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.5 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 1.7 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.7 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.7 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 57.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 2.7 | 10.6 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.6 | 7.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.6 | 4.7 | GO:0097636 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 1.5 | 4.5 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.4 | 5.4 | GO:0036019 | endolysosome(GO:0036019) |
| 1.1 | 11.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 1.1 | 8.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 1.0 | 4.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 1.0 | 10.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.9 | 2.7 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.8 | 8.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.7 | 4.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.7 | 2.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.7 | 7.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 3.3 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.6 | 2.9 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.6 | 4.6 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.5 | 11.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.5 | 2.1 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.5 | 3.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.5 | 3.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.5 | 1.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.5 | 1.8 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 9.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.4 | 1.2 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.4 | 18.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 1.5 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.4 | 2.7 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.4 | 5.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 1.9 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.3 | 1.0 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 0.3 | 3.7 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 3.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 2.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 4.5 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.3 | 5.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 5.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.3 | 5.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.0 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.2 | 9.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.2 | 2.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 1.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.2 | 1.7 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.2 | 1.5 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 2.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 18.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 29.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 4.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.2 | 3.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 0.5 | GO:0044393 | microspike(GO:0044393) |
| 0.2 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.2 | 1.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 12.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 4.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.2 | 2.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 2.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 4.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 13.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 37.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 1.0 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 2.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.0 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 2.3 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.1 | 2.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 4.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 4.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 2.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 1.1 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.5 | GO:1902710 | GABA receptor complex(GO:1902710) |
| 0.1 | 0.5 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 1.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 0.4 | GO:0071144 | activin responsive factor complex(GO:0032444) SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 11.6 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.1 | 9.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.8 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 2.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.9 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 21.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.1 | 7.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 4.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.7 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 1.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 6.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 3.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 6.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 2.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 4.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 47.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 2.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.2 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.7 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 1.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.9 | 55.5 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 3.5 | 10.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 3.3 | 20.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 2.8 | 8.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 2.7 | 10.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 2.7 | 8.0 | GO:0031403 | lithium ion binding(GO:0031403) |
| 2.5 | 7.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 2.4 | 12.0 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 2.2 | 6.6 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 2.2 | 6.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 2.1 | 6.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.8 | 5.5 | GO:0005186 | pheromone activity(GO:0005186) |
| 1.8 | 5.4 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 1.7 | 10.3 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 1.7 | 6.8 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 1.6 | 4.9 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 1.6 | 6.3 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 1.5 | 14.7 | GO:0035473 | lipase binding(GO:0035473) |
| 1.5 | 4.4 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 1.4 | 4.2 | GO:0015182 | L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.4 | 5.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.3 | 4.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.2 | 3.5 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 1.1 | 6.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 1.1 | 3.3 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.1 | 3.3 | GO:0038052 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 1.1 | 5.5 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) |
| 1.1 | 5.4 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 1.0 | 4.1 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 1.0 | 2.9 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 1.0 | 2.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.8 | 7.9 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.8 | 3.9 | GO:0016726 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.8 | 3.9 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.8 | 5.4 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.8 | 6.8 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.8 | 4.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.7 | 4.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 5.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 2.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.7 | 3.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.7 | 4.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.7 | 2.0 | GO:0050051 | leukotriene-B4 20-monooxygenase activity(GO:0050051) |
| 0.7 | 2.0 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.6 | 2.5 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 9.5 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.6 | 4.4 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.6 | 2.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.6 | 3.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.6 | 3.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.6 | 7.0 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.6 | 2.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.6 | 13.6 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.6 | 3.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.6 | 5.0 | GO:0046790 | virion binding(GO:0046790) |
| 0.5 | 5.5 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 3.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.5 | 2.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.5 | 3.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.5 | 1.6 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.5 | 2.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.5 | 4.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.5 | 3.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 3.5 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.5 | 1.4 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.5 | 3.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 3.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 7.9 | GO:0001848 | complement binding(GO:0001848) |
| 0.4 | 2.6 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 10.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.4 | 1.7 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.4 | 1.3 | GO:0031686 | uridine nucleotide receptor activity(GO:0015065) A1 adenosine receptor binding(GO:0031686) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.4 | 1.7 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.4 | 9.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 2.8 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.4 | 2.0 | GO:0070404 | NADH binding(GO:0070404) |
| 0.4 | 1.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.4 | 3.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 5.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.4 | 1.2 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.4 | 3.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 1.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.4 | 2.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 5.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 2.4 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.3 | 1.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.3 | 1.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.3 | 4.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.3 | 12.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.3 | 2.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 0.9 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.3 | 0.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.3 | 2.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 2.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 1.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 6.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.3 | 1.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 0.8 | GO:0017099 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 1.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 8.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.3 | 7.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.3 | 1.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.3 | 2.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.3 | 1.0 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 2.8 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.7 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.2 | 1.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.2 | 2.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.2 | 5.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 2.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 1.1 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 11.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.2 | 1.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 0.6 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.2 | 1.7 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 5.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.2 | 8.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.4 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.2 | 1.8 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.2 | 4.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.6 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 1.0 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 0.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.2 | 4.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 8.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.2 | 1.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 2.6 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.2 | 0.9 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 9.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 4.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.0 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 0.5 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.2 | 0.7 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 2.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 1.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 2.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 7.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 10.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.2 | 0.3 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.2 | 3.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 9.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 0.7 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.1 | 5.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.1 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.4 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.1 | 2.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.1 | 3.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.9 | GO:0001595 | angiotensin receptor activity(GO:0001595) |
| 0.1 | 0.9 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 1.6 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 3.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.6 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 5.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 2.2 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.6 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.7 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 11.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.5 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 1.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 4.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.6 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 1.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.5 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.9 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.7 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 3.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.9 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 4.1 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 2.3 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 1.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 3.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 1.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.7 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.1 | 13.8 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.1 | 1.1 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.8 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 2.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 1.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 3.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.6 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.1 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.6 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.8 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 1.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 3.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.8 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.1 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.0 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.0 | 0.9 | GO:0016853 | isomerase activity(GO:0016853) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 3.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 3.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 12.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.2 | 3.8 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 3.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 2.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 4.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 11.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.0 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.1 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 2.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 3.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.6 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 2.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 6.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 5.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 69.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.2 | 16.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.0 | 26.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.9 | 15.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.8 | 10.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.8 | 10.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.8 | 10.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.7 | 2.9 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.7 | 12.5 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.6 | 12.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.6 | 44.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 3.9 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.5 | 11.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.5 | 6.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.5 | 6.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.4 | 6.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.4 | 2.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.3 | 1.3 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.3 | 5.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.3 | 4.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.3 | 5.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 4.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 19.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.3 | 2.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 3.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 4.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.3 | 5.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 2.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 4.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 20.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 3.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 4.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 3.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 3.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 1.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.2 | 1.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.2 | 2.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 5.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 2.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 11.5 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 7.7 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.1 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.8 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |
| 0.1 | 1.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.0 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.1 | 6.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.7 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.7 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.1 | 0.6 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.1 | 3.5 | REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | Genes involved in Gastrin-CREB signalling pathway via PKC and MAPK |
| 0.1 | 1.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 9.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 2.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 5.0 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 1.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 5.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 0.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.5 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.8 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 2.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |