Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr2e1
Z-value: 7.31
Transcription factors associated with Nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr2e1
|
ENSMUSG00000019803.5 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr2e1 | mm10_v2_chr10_-_42583628_42583639 | 0.02 | 9.0e-01 | Click! |
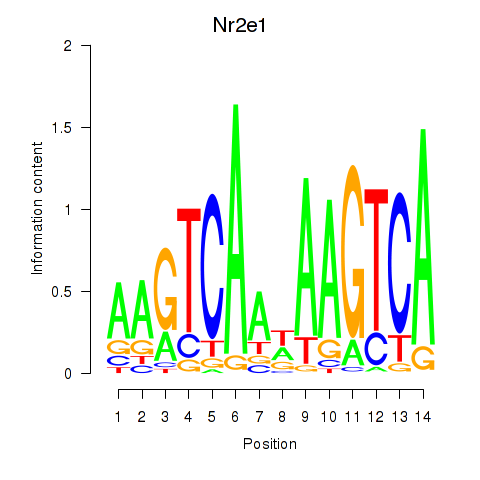
Activity profile of Nr2e1 motif
Sorted Z-values of Nr2e1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_60582152 | 112.42 |
ENSMUST00000098047.2
|
Mup10
|
major urinary protein 10 |
| chr4_-_62087261 | 110.58 |
ENSMUST00000107488.3
ENSMUST00000107472.1 ENSMUST00000084531.4 |
Mup3
|
major urinary protein 3 |
| chr4_-_60421933 | 108.24 |
ENSMUST00000107506.2
ENSMUST00000122381.1 ENSMUST00000118759.1 ENSMUST00000122177.1 |
Mup9
|
major urinary protein 9 |
| chr4_-_61303998 | 108.20 |
ENSMUST00000071005.8
ENSMUST00000075206.5 |
Mup14
|
major urinary protein 14 |
| chr4_-_60501903 | 106.90 |
ENSMUST00000084548.4
ENSMUST00000103012.3 ENSMUST00000107499.3 |
Mup1
|
major urinary protein 1 |
| chr4_-_61303802 | 102.87 |
ENSMUST00000125461.1
|
Mup14
|
major urinary protein 14 |
| chr4_-_60741275 | 101.16 |
ENSMUST00000117932.1
|
Mup12
|
major urinary protein 12 |
| chr4_-_61674094 | 91.86 |
ENSMUST00000098040.3
|
Mup18
|
major urinary protein 18 |
| chr4_-_60222580 | 84.70 |
ENSMUST00000095058.4
ENSMUST00000163931.1 |
Mup8
|
major urinary protein 8 |
| chr4_-_60662358 | 80.45 |
ENSMUST00000084544.4
ENSMUST00000098046.3 |
Mup11
|
major urinary protein 11 |
| chr4_-_60139857 | 78.54 |
ENSMUST00000107490.4
ENSMUST00000074700.2 |
Mup2
|
major urinary protein 2 |
| chr4_-_62054112 | 71.38 |
ENSMUST00000074018.3
|
Mup20
|
major urinary protein 20 |
| chr4_-_62150810 | 69.68 |
ENSMUST00000077719.3
|
Mup21
|
major urinary protein 21 |
| chr4_+_60003438 | 47.56 |
ENSMUST00000107517.1
ENSMUST00000107520.1 |
Mup6
|
major urinary protein 6 |
| chr6_-_141946791 | 45.38 |
ENSMUST00000168119.1
|
Slco1a1
|
solute carrier organic anion transporter family, member 1a1 |
| chr4_-_59960659 | 33.45 |
ENSMUST00000075973.2
|
Mup4
|
major urinary protein 4 |
| chr8_+_36489191 | 22.49 |
ENSMUST00000171777.1
|
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr14_+_29018205 | 19.91 |
ENSMUST00000055662.2
|
Lrtm1
|
leucine-rich repeats and transmembrane domains 1 |
| chr13_+_23839445 | 16.76 |
ENSMUST00000091698.4
ENSMUST00000110422.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr13_+_23839401 | 16.04 |
ENSMUST00000039721.7
ENSMUST00000166467.1 |
Slc17a3
|
solute carrier family 17 (sodium phosphate), member 3 |
| chr14_+_32028989 | 15.83 |
ENSMUST00000022460.4
|
Galnt15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr5_-_87140318 | 15.82 |
ENSMUST00000067790.6
ENSMUST00000113327.1 |
Ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr2_-_73386396 | 15.63 |
ENSMUST00000112044.1
ENSMUST00000112043.1 ENSMUST00000076463.5 |
Gpr155
|
G protein-coupled receptor 155 |
| chr5_-_87424201 | 14.28 |
ENSMUST00000072818.5
|
Ugt2b38
|
UDP glucuronosyltransferase 2 family, polypeptide B38 |
| chr4_-_119415494 | 14.02 |
ENSMUST00000063642.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr5_+_87000838 | 12.18 |
ENSMUST00000031186.7
|
Ugt2b35
|
UDP glucuronosyltransferase 2 family, polypeptide B35 |
| chr17_+_32685610 | 11.56 |
ENSMUST00000168171.1
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr17_+_32685655 | 11.31 |
ENSMUST00000008801.6
|
Cyp4f15
|
cytochrome P450, family 4, subfamily f, polypeptide 15 |
| chr5_-_86906937 | 9.55 |
ENSMUST00000031181.9
ENSMUST00000113333.1 |
Ugt2b34
|
UDP glucuronosyltransferase 2 family, polypeptide B34 |
| chr1_+_24678536 | 9.12 |
ENSMUST00000095062.3
|
Lmbrd1
|
LMBR1 domain containing 1 |
| chr5_-_88527841 | 9.11 |
ENSMUST00000087033.3
|
Igj
|
immunoglobulin joining chain |
| chr7_+_114768736 | 8.94 |
ENSMUST00000117543.1
|
Insc
|
inscuteable homolog (Drosophila) |
| chr7_+_114768767 | 7.86 |
ENSMUST00000151464.1
|
Insc
|
inscuteable homolog (Drosophila) |
| chr7_-_80405425 | 7.84 |
ENSMUST00000107362.3
ENSMUST00000135306.1 |
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr3_+_96525163 | 7.62 |
ENSMUST00000049208.9
|
Hfe2
|
hemochromatosis type 2 (juvenile) (human homolog) |
| chr15_+_41830921 | 6.90 |
ENSMUST00000166917.1
|
Oxr1
|
oxidation resistance 1 |
| chr9_-_71163224 | 6.80 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chrM_+_7005 | 6.54 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr7_+_107209439 | 5.84 |
ENSMUST00000098135.1
|
Rbmxl2
|
RNA binding motif protein, X-linked-like 2 |
| chr3_-_146781351 | 5.42 |
ENSMUST00000005164.7
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr12_+_87147703 | 5.35 |
ENSMUST00000063117.8
|
Gstz1
|
glutathione transferase zeta 1 (maleylacetoacetate isomerase) |
| chr17_+_33919332 | 4.77 |
ENSMUST00000025161.7
|
Tapbp
|
TAP binding protein |
| chr8_-_61902669 | 4.09 |
ENSMUST00000121785.1
ENSMUST00000034057.7 |
Palld
|
palladin, cytoskeletal associated protein |
| chr12_+_111166413 | 3.95 |
ENSMUST00000021706.4
|
Traf3
|
TNF receptor-associated factor 3 |
| chr9_-_42457594 | 3.81 |
ENSMUST00000125995.1
|
Tbcel
|
tubulin folding cofactor E-like |
| chr6_+_65590382 | 3.63 |
ENSMUST00000114236.1
|
Tnip3
|
TNFAIP3 interacting protein 3 |
| chr12_-_87147883 | 3.16 |
ENSMUST00000037788.4
|
Pomt2
|
protein-O-mannosyltransferase 2 |
| chr3_+_14578609 | 2.70 |
ENSMUST00000029069.6
ENSMUST00000165922.2 |
E2f5
|
E2F transcription factor 5 |
| chrX_-_95658416 | 2.69 |
ENSMUST00000044382.6
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr12_+_86734381 | 2.60 |
ENSMUST00000095527.5
|
Gm6772
|
predicted gene 6772 |
| chr5_-_129704259 | 2.53 |
ENSMUST00000042266.6
|
Sept14
|
septin 14 |
| chr10_+_18469958 | 2.48 |
ENSMUST00000162891.1
ENSMUST00000100054.3 |
Nhsl1
|
NHS-like 1 |
| chr13_+_4771649 | 2.44 |
ENSMUST00000065956.3
|
Gm5444
|
predicted gene 5444 |
| chr2_-_23040177 | 2.42 |
ENSMUST00000114544.3
ENSMUST00000139038.1 ENSMUST00000126112.1 ENSMUST00000178908.1 ENSMUST00000078977.7 ENSMUST00000140164.1 ENSMUST00000149719.1 |
Abi1
|
abl-interactor 1 |
| chr2_-_23040137 | 2.42 |
ENSMUST00000091394.6
ENSMUST00000093171.6 |
Abi1
|
abl-interactor 1 |
| chr12_+_111166536 | 2.35 |
ENSMUST00000060274.6
|
Traf3
|
TNF receptor-associated factor 3 |
| chr10_-_24092320 | 2.25 |
ENSMUST00000092654.2
|
Taar8b
|
trace amine-associated receptor 8B |
| chr2_-_23040092 | 2.18 |
ENSMUST00000153931.1
ENSMUST00000123948.1 |
Abi1
|
abl-interactor 1 |
| chr14_+_26693267 | 2.16 |
ENSMUST00000022433.4
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chrY_+_23097865 | 1.70 |
ENSMUST00000179975.1
|
Gm21661
|
predicted gene, 21661 |
| chrY_-_74789646 | 1.70 |
ENSMUST00000179145.1
|
Gm21076
|
predicted gene, 21076 |
| chrY_-_75466372 | 1.70 |
ENSMUST00000177869.1
|
Gm21922
|
predicted gene, 21922 |
| chrY_-_83715782 | 1.70 |
ENSMUST00000180018.1
|
Gm21741
|
predicted gene, 21741 |
| chrY_-_83853059 | 1.70 |
ENSMUST00000180274.1
|
Gm20863
|
predicted gene, 20863 |
| chrY_-_83288579 | 1.70 |
ENSMUST00000178222.1
|
Gm21864
|
predicted gene, 21864 |
| chrY_-_74951338 | 1.69 |
ENSMUST00000178873.1
|
Gm21869
|
predicted gene, 21869 |
| chrY_+_38247727 | 1.69 |
ENSMUST00000178740.1
|
Gm21735
|
predicted gene, 21735 |
| chrY_-_85454516 | 1.66 |
ENSMUST00000178264.1
|
Gm21785
|
predicted gene, 21785 |
| chrY_-_85814909 | 1.66 |
ENSMUST00000177729.1
|
Gm21881
|
predicted gene, 21881 |
| chrY_+_34561734 | 1.65 |
ENSMUST00000179779.1
|
Gm21201
|
predicted gene, 21201 |
| chrY_-_78762038 | 1.65 |
ENSMUST00000179381.1
|
Gm20808
|
predicted gene, 20808 |
| chrY_-_80682503 | 1.65 |
ENSMUST00000179632.1
|
Gm20791
|
predicted gene, 20791 |
| chrY_-_82787743 | 1.65 |
ENSMUST00000180145.1
|
Gm21344
|
predicted gene, 21344 |
| chrY_+_68464868 | 1.63 |
ENSMUST00000179830.1
|
Gm21127
|
predicted gene, 21127 |
| chrY_-_77149422 | 1.63 |
ENSMUST00000178718.1
|
Gm20859
|
predicted gene, 20859 |
| chrY_+_35301254 | 1.63 |
ENSMUST00000179413.1
|
Gm20799
|
predicted gene, 20799 |
| chrY_-_52262626 | 1.63 |
ENSMUST00000178928.1
|
Gm21245
|
predicted gene, 21245 |
| chrY_-_64859352 | 1.63 |
ENSMUST00000178067.1
|
Gm20921
|
predicted gene, 20921 |
| chrY_-_72039692 | 1.63 |
ENSMUST00000178109.1
|
Gm20842
|
predicted gene, 20842 |
| chrY_+_5727496 | 1.61 |
ENSMUST00000180372.1
|
Gm20914
|
predicted gene, 20914 |
| chrY_-_82020055 | 1.61 |
ENSMUST00000177769.1
|
Gm21790
|
predicted gene, 21790 |
| chrY_-_50955531 | 1.59 |
ENSMUST00000179102.1
|
Gm20887
|
predicted gene, 20887 |
| chrY_-_82862028 | 1.59 |
ENSMUST00000178638.1
|
Gm20849
|
predicted gene, 20849 |
| chrY_+_23381504 | 1.57 |
ENSMUST00000180340.1
|
Gm20926
|
predicted gene, 20926 |
| chrY_+_24381817 | 1.57 |
ENSMUST00000177717.1
|
Gm21845
|
predicted gene, 21845 |
| chrY_+_24456582 | 1.57 |
ENSMUST00000179663.1
|
Gm20809
|
predicted gene, 20809 |
| chrY_+_24894925 | 1.57 |
ENSMUST00000177807.1
|
Gm21350
|
predicted gene, 21350 |
| chrY_+_24969606 | 1.57 |
ENSMUST00000179822.1
|
Gm21412
|
predicted gene, 21412 |
| chrY_+_25408061 | 1.57 |
ENSMUST00000179773.1
|
Gm21791
|
predicted gene, 21791 |
| chrY_+_25482701 | 1.57 |
ENSMUST00000179838.1
|
Gm21427
|
predicted gene, 21427 |
| chrY_+_25921142 | 1.57 |
ENSMUST00000178189.1
|
Gm21890
|
predicted gene, 21890 |
| chrY_+_26434218 | 1.57 |
ENSMUST00000178570.1
|
Gm21755
|
predicted gene, 21755 |
| chrY_+_27179288 | 1.57 |
ENSMUST00000179877.1
|
Gm21771
|
predicted gene, 21771 |
| chrY_+_27924318 | 1.57 |
ENSMUST00000180279.1
|
Gm20891
|
predicted gene, 20891 |
| chrY_+_28443743 | 1.57 |
ENSMUST00000179068.1
|
Gm21642
|
predicted gene, 21642 |
| chrY_+_35644173 | 1.57 |
ENSMUST00000178709.1
|
Gm20798
|
predicted gene, 20798 |
| chrY_+_68539472 | 1.57 |
ENSMUST00000177765.1
|
Gm20816
|
predicted gene, 20816 |
| chrY_-_51606491 | 1.57 |
ENSMUST00000178269.1
|
Gm21387
|
predicted gene, 21387 |
| chrY_-_53051486 | 1.57 |
ENSMUST00000177852.1
|
Gm21268
|
predicted gene, 21268 |
| chrY_-_53342254 | 1.57 |
ENSMUST00000177621.1
|
Gm21285
|
predicted gene, 21285 |
| chrY_-_64227755 | 1.57 |
ENSMUST00000177938.1
|
Gm20882
|
predicted gene, 20882 |
| chrY_-_65649184 | 1.57 |
ENSMUST00000178339.1
|
Gm20936
|
predicted gene, 20936 |
| chrY_-_65939962 | 1.57 |
ENSMUST00000180295.1
|
Gm20933
|
predicted gene, 20933 |
| chrY_-_72383262 | 1.57 |
ENSMUST00000180348.1
|
Gm20844
|
predicted gene, 20844 |
| chrY_-_72820330 | 1.57 |
ENSMUST00000179517.1
|
Gm20846
|
predicted gene, 20846 |
| chrY_-_73239904 | 1.57 |
ENSMUST00000179537.1
|
Gm20800
|
predicted gene, 20800 |
| chrY_-_77074728 | 1.57 |
ENSMUST00000177623.1
|
Gm21180
|
predicted gene, 21180 |
| chrY_-_81204493 | 1.57 |
ENSMUST00000179373.1
|
Gm21282
|
predicted gene, 21282 |
| chrY_-_81725302 | 1.57 |
ENSMUST00000177798.1
|
Gm20912
|
predicted gene, 20912 |
| chrY_-_49318753 | 1.54 |
ENSMUST00000177930.1
|
Gm21308
|
predicted gene, 21308 |
| chrY_-_61946433 | 1.54 |
ENSMUST00000178579.1
|
Gm20803
|
predicted gene, 20803 |
| chrY_-_63501598 | 1.54 |
ENSMUST00000177673.1
|
Gm21634
|
predicted gene, 21634 |
| chrY_-_70197533 | 1.54 |
ENSMUST00000178860.1
|
Gm21672
|
predicted gene, 21672 |
| chrY_-_77961690 | 1.54 |
ENSMUST00000178900.1
|
Gm20867
|
predicted gene, 20867 |
| chr6_+_42400238 | 1.48 |
ENSMUST00000057398.3
|
Tas2r143
|
taste receptor, type 2, member 143 |
| chrX_-_95658379 | 1.47 |
ENSMUST00000119640.1
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chrY_+_68023542 | 1.46 |
ENSMUST00000178448.1
|
Gm20935
|
predicted gene, 20935 |
| chrY_-_49843610 | 1.46 |
ENSMUST00000177618.1
|
Gm21316
|
predicted gene, 21316 |
| chrY_-_52608321 | 1.46 |
ENSMUST00000179559.1
|
Gm21249
|
predicted gene, 21249 |
| chrY_-_65206008 | 1.46 |
ENSMUST00000177663.1
|
Gm20924
|
predicted gene, 20924 |
| chrY_-_70706829 | 1.46 |
ENSMUST00000179571.1
|
Gm20886
|
predicted gene, 20886 |
| chr4_+_135728116 | 1.45 |
ENSMUST00000102546.3
|
Il22ra1
|
interleukin 22 receptor, alpha 1 |
| chrY_+_37100588 | 1.43 |
ENSMUST00000180349.1
|
Gm21799
|
predicted gene, 21799 |
| chrY_-_86944319 | 1.41 |
ENSMUST00000178548.1
|
Gm21470
|
predicted gene, 21470 |
| chrY_-_86870055 | 1.39 |
ENSMUST00000178151.1
|
Gm21462
|
predicted gene, 21462 |
| chrY_-_54415539 | 1.38 |
ENSMUST00000178413.1
|
Gm21740
|
predicted gene, 21740 |
| chrY_-_55484007 | 1.38 |
ENSMUST00000178346.1
|
Gm21733
|
predicted gene, 21733 |
| chrY_-_56250174 | 1.38 |
ENSMUST00000179016.1
|
Gm21851
|
predicted gene, 21851 |
| chrY_-_57457990 | 1.38 |
ENSMUST00000178649.1
|
Gm21868
|
predicted gene, 21868 |
| chrY_+_28733498 | 1.35 |
ENSMUST00000180127.1
|
Gm20810
|
predicted gene, 20810 |
| chrY_+_38172994 | 1.33 |
ENSMUST00000178866.1
|
Gm21924
|
predicted gene, 21924 |
| chrY_+_23031235 | 1.28 |
ENSMUST00000178910.1
|
Gm21469
|
predicted gene, 21469 |
| chrY_-_54186670 | 1.28 |
ENSMUST00000178094.1
|
Gm21302
|
predicted gene, 21302 |
| chrY_-_55044004 | 1.28 |
ENSMUST00000179739.1
|
Gm20930
|
predicted gene, 20930 |
| chrY_-_57017520 | 1.28 |
ENSMUST00000179148.1
|
Gm20913
|
predicted gene, 20913 |
| chrY_-_66740610 | 1.28 |
ENSMUST00000178761.1
|
Gm20852
|
predicted gene, 20852 |
| chrY_-_73536230 | 1.28 |
ENSMUST00000180169.1
|
Gm20902
|
predicted gene, 20902 |
| chrY_-_75026075 | 1.28 |
ENSMUST00000177922.1
|
Gm20813
|
predicted gene, 20813 |
| chrY_-_75540398 | 1.28 |
ENSMUST00000178387.1
|
Gm21111
|
predicted gene, 21111 |
| chrY_-_76061651 | 1.28 |
ENSMUST00000179698.1
|
Gm20904
|
predicted gene, 20904 |
| chrY_-_83790566 | 1.28 |
ENSMUST00000178994.1
|
Gm20862
|
predicted gene, 20862 |
| chrY_-_83927735 | 1.28 |
ENSMUST00000179616.1
|
Gm20864
|
predicted gene, 20864 |
| chrY_-_84440964 | 1.28 |
ENSMUST00000179959.1
|
Gm21380
|
predicted gene, 21380 |
| chrY_-_85889645 | 1.28 |
ENSMUST00000177993.1
|
Gm21435
|
predicted gene, 21435 |
| chrY_+_21166083 | 1.28 |
ENSMUST00000178234.1
|
Gm20909
|
predicted gene, 20909 |
| chrY_+_23306767 | 1.28 |
ENSMUST00000178648.1
|
Gm20925
|
predicted gene, 20925 |
| chrY_-_53416775 | 1.28 |
ENSMUST00000179137.1
|
Gm20747
|
predicted gene, 20747 |
| chrY_-_73314512 | 1.28 |
ENSMUST00000177855.1
|
Gm21683
|
predicted gene, 21683 |
| chrY_-_81799967 | 1.28 |
ENSMUST00000179785.1
|
Gm20801
|
predicted gene, 20801 |
| chrY_-_83362381 | 1.28 |
ENSMUST00000179849.1
|
Gm21802
|
predicted gene, 21802 |
| chrY_-_86434894 | 1.28 |
ENSMUST00000179044.1
|
Gm20860
|
predicted gene, 20860 |
| chrY_-_79590982 | 1.28 |
ENSMUST00000179173.1
|
Gm21823
|
predicted gene, 21823 |
| chrY_-_88805474 | 1.28 |
ENSMUST00000178351.1
|
Gm21287
|
predicted gene, 21287 |
| chrY_+_26508824 | 1.25 |
ENSMUST00000179045.1
|
Gm20895
|
predicted gene, 20895 |
| chrY_+_26947544 | 1.25 |
ENSMUST00000180115.1
|
Gm20892
|
predicted gene, 20892 |
| chrY_+_27253916 | 1.25 |
ENSMUST00000179896.1
|
Gm21476
|
predicted gene, 21476 |
| chrY_+_27692628 | 1.25 |
ENSMUST00000178381.1
|
Gm21506
|
predicted gene, 21506 |
| chrY_+_27998840 | 1.25 |
ENSMUST00000180057.1
|
Gm20893
|
predicted gene, 20893 |
| chrY_+_28518281 | 1.25 |
ENSMUST00000178131.1
|
Gm21804
|
predicted gene, 21804 |
| chrY_-_50435859 | 1.25 |
ENSMUST00000178299.1
|
Gm20881
|
predicted gene, 20881 |
| chrY_-_51681223 | 1.25 |
ENSMUST00000179651.1
|
Gm21396
|
predicted gene, 21396 |
| chrY_-_63055872 | 1.25 |
ENSMUST00000177788.1
|
Gm21617
|
predicted gene, 21617 |
| chrY_-_64302598 | 1.25 |
ENSMUST00000179622.1
|
Gm20907
|
predicted gene, 20907 |
| chrY_-_50361155 | 1.25 |
ENSMUST00000178141.1
|
Gm20880
|
predicted gene, 20880 |
| chrY_-_62981165 | 1.25 |
ENSMUST00000178521.1
|
Gm21530
|
predicted gene, 21530 |
| chrY_-_71224005 | 1.25 |
ENSMUST00000177571.1
|
Gm20866
|
predicted gene, 20866 |
| chrY_+_22513860 | 1.24 |
ENSMUST00000178985.1
|
Gm21065
|
predicted gene, 21065 |
| chrY_+_34486834 | 1.24 |
ENSMUST00000178803.1
|
Gm21184
|
predicted gene, 21184 |
| chrY_+_34781126 | 1.24 |
ENSMUST00000179452.1
|
Gm20853
|
predicted gene, 20853 |
| chrY_+_46757399 | 1.24 |
ENSMUST00000179950.1
|
Gm21247
|
predicted gene, 21247 |
| chrY_+_69017824 | 1.24 |
ENSMUST00000179438.1
|
Gm20818
|
predicted gene, 20818 |
| chrY_+_69352610 | 1.24 |
ENSMUST00000178496.1
|
Gm21160
|
predicted gene, 21160 |
| chrY_-_9345654 | 1.24 |
ENSMUST00000179590.1
|
Gm21812
|
predicted gene, 21812 |
| chrY_-_48670459 | 1.24 |
ENSMUST00000179582.1
|
Gm21281
|
predicted gene, 21281 |
| chrY_-_54489811 | 1.24 |
ENSMUST00000177749.1
|
Gm21912
|
predicted gene, 21912 |
| chrY_-_55558343 | 1.24 |
ENSMUST00000178916.1
|
Gm21330
|
predicted gene, 21330 |
| chrY_-_56324320 | 1.24 |
ENSMUST00000177558.1
|
Gm21819
|
predicted gene, 21819 |
| chrY_-_61460715 | 1.24 |
ENSMUST00000178176.1
|
Gm21573
|
predicted gene, 21573 |
| chrY_-_67046566 | 1.24 |
ENSMUST00000179136.1
|
Ssty2
|
spermiogenesis specific transcript on the Y 2 |
| chrY_-_74300895 | 1.24 |
ENSMUST00000178984.1
|
Gm20805
|
predicted gene, 20805 |
| chrY_-_76576410 | 1.24 |
ENSMUST00000179497.1
|
Gm21163
|
predicted gene, 21163 |
| chrY_-_80463929 | 1.24 |
ENSMUST00000180185.1
|
Gm21257
|
predicted gene, 21257 |
| chrY_-_82568207 | 1.24 |
ENSMUST00000178088.1
|
Gm21333
|
predicted gene, 21333 |
| chrY_+_35569461 | 1.23 |
ENSMUST00000180002.1
|
Gm21872
|
predicted gene, 21872 |
| chrY_+_37884615 | 1.22 |
ENSMUST00000178736.1
|
Gm20922
|
predicted gene, 20922 |
| chr12_+_111166349 | 1.21 |
ENSMUST00000117269.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chrX_-_134111708 | 1.21 |
ENSMUST00000159259.1
ENSMUST00000113275.3 |
Nox1
|
NADPH oxidase 1 |
| chrY_-_50880880 | 1.20 |
ENSMUST00000178018.1
|
Gm20889
|
predicted gene, 20889 |
| chrY_-_85529518 | 1.17 |
ENSMUST00000178889.1
|
Gm20854
|
predicted gene, 20854 |
| chrY_-_79074940 | 1.14 |
ENSMUST00000178606.1
|
Gm20919
|
predicted gene, 20919 |
| chrX_-_95658392 | 1.07 |
ENSMUST00000120620.1
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chrY_-_79149787 | 1.01 |
ENSMUST00000179922.1
|
Gm20917
|
predicted gene, 20917 |
| chrY_-_79959257 | 1.00 |
ENSMUST00000178326.1
|
Gm21248
|
predicted gene, 21248 |
| chrY_+_37430489 | 0.98 |
ENSMUST00000177751.1
|
Gm21835
|
predicted gene, 21835 |
| chrY_-_78836543 | 0.98 |
ENSMUST00000180324.1
|
Gm20806
|
predicted gene, 20806 |
| chrY_-_80757034 | 0.98 |
ENSMUST00000180158.1
|
Gm20841
|
predicted gene, 20841 |
| chrY_-_9132561 | 0.94 |
ENSMUST00000171947.2
|
Gm21292
|
predicted gene, 21292 |
| chrY_-_78035714 | 0.92 |
ENSMUST00000178749.1
|
Gm21658
|
predicted gene, 21658 |
| chr15_-_97705787 | 0.77 |
ENSMUST00000023104.5
|
Rpap3
|
RNA polymerase II associated protein 3 |
| chrY_-_84489780 | 0.64 |
ENSMUST00000179727.1
|
Gm21782
|
predicted gene, 21782 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr2e1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.0 | 187.4 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 11.9 | 71.4 | GO:0008355 | olfactory learning(GO:0008355) |
| 5.8 | 156.0 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 2.3 | 32.8 | GO:0015747 | urate transport(GO:0015747) |
| 2.0 | 22.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 2.0 | 7.8 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 1.7 | 6.8 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 1.1 | 5.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 1.1 | 3.2 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.7 | 4.8 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.6 | 6.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.6 | 6.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.5 | 5.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.4 | 7.6 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.4 | 1.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.4 | 16.8 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.4 | 3.6 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.3 | 9.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.2 | 2.5 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 19.9 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 7.0 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.2 | 4.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 2.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 6.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.3 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 1.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 12.7 | GO:0050890 | cognition(GO:0050890) |
| 0.0 | 1.4 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 2.7 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.6 | 4.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.5 | 7.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.5 | 7.8 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.4 | 5.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.4 | 6.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.3 | 6.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 9.1 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 1.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 2.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 52.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 303.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 49.6 | GO:0045177 | apical part of cell(GO:0045177) |
| 0.1 | 4.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 15.8 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 15.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.7 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 57.8 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 59.4 | 178.3 | GO:0005186 | pheromone activity(GO:0005186) |
| 6.0 | 144.0 | GO:0005550 | pheromone binding(GO:0005550) |
| 4.1 | 32.8 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 3.7 | 22.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 1.4 | 51.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 1.4 | 6.8 | GO:0015254 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 1.1 | 40.9 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 1.0 | 9.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.8 | 15.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.7 | 7.6 | GO:0036122 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.7 | 4.8 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.5 | 5.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 3.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.5 | 7.8 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.5 | 1.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.5 | 1.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.5 | 7.0 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.4 | 6.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.2 | 1.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.2 | 6.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 22.9 | GO:0046906 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.1 | 19.9 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 5.3 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.1 | 16.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 2.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 3.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 12.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 7.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 6.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 7.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.8 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.6 | 7.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.4 | 6.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 5.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 5.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |