Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Nr0b1
Z-value: 1.28
Transcription factors associated with Nr0b1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nr0b1
|
ENSMUSG00000025056.4 | nuclear receptor subfamily 0, group B, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nr0b1 | mm10_v2_chrX_+_86191764_86191782 | -0.58 | 1.8e-04 | Click! |
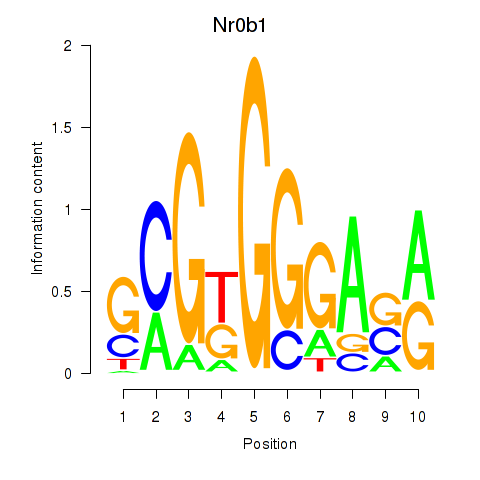
Activity profile of Nr0b1 motif
Sorted Z-values of Nr0b1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_88087802 | 5.33 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_-_57059795 | 3.99 |
ENSMUST00000040280.7
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr16_-_20621255 | 2.78 |
ENSMUST00000052939.2
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr4_+_148130883 | 2.29 |
ENSMUST00000084129.2
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr7_+_28180226 | 2.23 |
ENSMUST00000172467.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr7_+_28180272 | 2.18 |
ENSMUST00000173223.1
|
Dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1b |
| chr16_+_44173239 | 2.15 |
ENSMUST00000119746.1
|
Gm608
|
predicted gene 608 |
| chr13_-_29984219 | 1.98 |
ENSMUST00000146092.1
|
E2f3
|
E2F transcription factor 3 |
| chr9_+_80165013 | 1.63 |
ENSMUST00000035889.8
ENSMUST00000113268.1 |
Myo6
|
myosin VI |
| chr9_+_80165079 | 1.62 |
ENSMUST00000184480.1
|
Myo6
|
myosin VI |
| chr6_+_134830145 | 1.57 |
ENSMUST00000046303.5
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr8_-_95434869 | 1.57 |
ENSMUST00000034249.6
|
Gtl3
|
gene trap locus 3 |
| chr11_+_94328242 | 1.53 |
ENSMUST00000021227.5
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_-_128589650 | 1.52 |
ENSMUST00000082059.6
|
Erbb3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) |
| chr2_+_157279065 | 1.49 |
ENSMUST00000029171.5
|
Rpn2
|
ribophorin II |
| chr10_+_116301374 | 1.47 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr10_-_127666673 | 1.41 |
ENSMUST00000026469.2
|
Nab2
|
Ngfi-A binding protein 2 |
| chr2_-_65238573 | 1.40 |
ENSMUST00000090896.3
ENSMUST00000155082.1 |
Cobll1
|
Cobl-like 1 |
| chr1_+_140246216 | 1.38 |
ENSMUST00000119786.1
ENSMUST00000120796.1 ENSMUST00000060201.8 ENSMUST00000120709.1 |
Kcnt2
|
potassium channel, subfamily T, member 2 |
| chr2_+_4718145 | 1.36 |
ENSMUST00000056914.6
|
Bend7
|
BEN domain containing 7 |
| chr16_-_45844228 | 1.30 |
ENSMUST00000076333.5
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr16_-_45844303 | 1.29 |
ENSMUST00000036355.6
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr2_+_59160884 | 1.29 |
ENSMUST00000037903.8
|
Pkp4
|
plakophilin 4 |
| chr2_+_157279026 | 1.28 |
ENSMUST00000116380.2
|
Rpn2
|
ribophorin II |
| chr6_+_134830216 | 1.27 |
ENSMUST00000111937.1
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr17_-_26508463 | 1.25 |
ENSMUST00000025025.6
|
Dusp1
|
dual specificity phosphatase 1 |
| chr4_-_118134869 | 1.17 |
ENSMUST00000097912.1
ENSMUST00000030263.2 ENSMUST00000106410.1 |
St3gal3
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3 |
| chr9_+_57521232 | 1.14 |
ENSMUST00000000090.6
|
Cox5a
|
cytochrome c oxidase subunit Va |
| chr17_-_23745829 | 1.11 |
ENSMUST00000046525.8
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr3_-_30509462 | 1.10 |
ENSMUST00000173899.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr11_+_94327984 | 1.07 |
ENSMUST00000107818.2
ENSMUST00000051221.6 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr8_+_71371283 | 1.03 |
ENSMUST00000110051.1
ENSMUST00000002469.8 ENSMUST00000110052.1 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr2_-_80581234 | 1.02 |
ENSMUST00000028386.5
|
Nckap1
|
NCK-associated protein 1 |
| chr2_-_65238625 | 1.01 |
ENSMUST00000112429.2
ENSMUST00000102726.1 ENSMUST00000112430.1 |
Cobll1
|
Cobl-like 1 |
| chr13_-_64274879 | 1.00 |
ENSMUST00000109770.1
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr2_-_80581380 | 0.97 |
ENSMUST00000111760.2
|
Nckap1
|
NCK-associated protein 1 |
| chr11_+_23306884 | 0.97 |
ENSMUST00000180046.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr3_-_139075178 | 0.97 |
ENSMUST00000098574.2
ENSMUST00000029796.6 |
Rap1gds1
|
RAP1, GTP-GDP dissociation stimulator 1 |
| chr8_+_55940453 | 0.95 |
ENSMUST00000000275.7
|
Glra3
|
glycine receptor, alpha 3 subunit |
| chr14_+_11553523 | 0.94 |
ENSMUST00000022264.6
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G |
| chr7_-_80371197 | 0.92 |
ENSMUST00000098346.3
|
Man2a2
|
mannosidase 2, alpha 2 |
| chr4_+_116221633 | 0.86 |
ENSMUST00000030464.7
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chrX_-_92016183 | 0.85 |
ENSMUST00000099471.2
ENSMUST00000072269.1 |
Mageb1
|
melanoma antigen, family B, 1 |
| chr5_+_121711609 | 0.84 |
ENSMUST00000051950.7
|
Atxn2
|
ataxin 2 |
| chr8_-_85080652 | 0.84 |
ENSMUST00000152785.1
|
Wdr83
|
WD repeat domain containing 83 |
| chr8_-_85080679 | 0.78 |
ENSMUST00000093357.5
|
Wdr83
|
WD repeat domain containing 83 |
| chr3_+_54156039 | 0.77 |
ENSMUST00000029311.6
|
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr18_+_35829798 | 0.77 |
ENSMUST00000060722.6
|
Cxxc5
|
CXXC finger 5 |
| chr13_-_31559333 | 0.75 |
ENSMUST00000170573.1
|
A530084C06Rik
|
RIKEN cDNA A530084C06 gene |
| chr17_-_70851189 | 0.74 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr10_+_39732099 | 0.73 |
ENSMUST00000019986.6
|
Rev3l
|
REV3-like, catalytic subunit of DNA polymerase zeta RAD54 like (S. cerevisiae) |
| chr3_-_107458895 | 0.70 |
ENSMUST00000009617.8
|
Kcnc4
|
potassium voltage gated channel, Shaw-related subfamily, member 4 |
| chr10_+_90829409 | 0.70 |
ENSMUST00000182202.1
ENSMUST00000182966.1 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_+_116221590 | 0.70 |
ENSMUST00000147292.1
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr10_-_81350389 | 0.67 |
ENSMUST00000020454.4
ENSMUST00000105324.2 ENSMUST00000154609.2 ENSMUST00000105323.1 |
Hmg20b
|
high mobility group 20B |
| chr17_-_80728026 | 0.66 |
ENSMUST00000112389.2
ENSMUST00000025089.7 |
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr3_+_86224665 | 0.66 |
ENSMUST00000107635.1
|
Lrba
|
LPS-responsive beige-like anchor |
| chr15_+_35371498 | 0.65 |
ENSMUST00000048646.7
|
Vps13b
|
vacuolar protein sorting 13B (yeast) |
| chr13_+_41249841 | 0.65 |
ENSMUST00000165561.2
|
Smim13
|
small integral membrane protein 13 |
| chr1_+_162570687 | 0.64 |
ENSMUST00000050010.4
ENSMUST00000150040.1 |
Vamp4
|
vesicle-associated membrane protein 4 |
| chr17_-_84790517 | 0.63 |
ENSMUST00000112308.2
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr5_-_99037035 | 0.63 |
ENSMUST00000031277.6
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr17_+_88530113 | 0.59 |
ENSMUST00000038551.6
|
Ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr4_-_138913915 | 0.57 |
ENSMUST00000097830.3
|
Otud3
|
OTU domain containing 3 |
| chr10_+_90829538 | 0.57 |
ENSMUST00000179694.2
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_47454320 | 0.54 |
ENSMUST00000031697.8
|
Cul1
|
cullin 1 |
| chr2_-_144331695 | 0.54 |
ENSMUST00000103171.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr11_-_70687917 | 0.54 |
ENSMUST00000108545.2
ENSMUST00000120261.1 ENSMUST00000036299.7 ENSMUST00000119120.1 ENSMUST00000100933.3 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr16_+_57121705 | 0.53 |
ENSMUST00000166897.1
|
Tomm70a
|
translocase of outer mitochondrial membrane 70 homolog A (yeast) |
| chr5_-_99037342 | 0.51 |
ENSMUST00000161490.1
|
Prkg2
|
protein kinase, cGMP-dependent, type II |
| chr4_-_123750236 | 0.48 |
ENSMUST00000102636.3
|
Akirin1
|
akirin 1 |
| chr10_+_127078886 | 0.48 |
ENSMUST00000039259.6
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr10_+_38965515 | 0.48 |
ENSMUST00000019992.5
|
Lama4
|
laminin, alpha 4 |
| chr4_-_135971894 | 0.45 |
ENSMUST00000105852.1
|
Lypla2
|
lysophospholipase 2 |
| chr11_+_105181527 | 0.43 |
ENSMUST00000106941.2
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr7_-_65371210 | 0.39 |
ENSMUST00000102592.3
|
Tjp1
|
tight junction protein 1 |
| chr6_-_52191695 | 0.38 |
ENSMUST00000101395.2
|
Hoxa4
|
homeobox A4 |
| chr5_+_110230975 | 0.38 |
ENSMUST00000031474.7
ENSMUST00000086674.5 |
Ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr15_-_38078842 | 0.38 |
ENSMUST00000110336.2
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr11_+_23306910 | 0.37 |
ENSMUST00000137823.1
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_+_105181793 | 0.37 |
ENSMUST00000092537.3
ENSMUST00000015107.6 ENSMUST00000145048.1 |
Tlk2
|
tousled-like kinase 2 (Arabidopsis) |
| chr13_+_94358943 | 0.35 |
ENSMUST00000022196.3
|
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr11_-_74723829 | 0.35 |
ENSMUST00000102520.2
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr4_-_11386394 | 0.34 |
ENSMUST00000155519.1
|
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr2_+_153842981 | 0.32 |
ENSMUST00000175856.1
|
Efcab8
|
EF-hand calcium binding domain 8 |
| chr5_-_31193105 | 0.30 |
ENSMUST00000166769.1
|
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr2_+_30066419 | 0.30 |
ENSMUST00000067996.6
|
Set
|
SET nuclear oncogene |
| chr1_+_59764264 | 0.28 |
ENSMUST00000087435.5
|
Bmpr2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr5_-_31193008 | 0.26 |
ENSMUST00000114605.1
ENSMUST00000114603.1 |
Eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr9_-_110117379 | 0.25 |
ENSMUST00000111991.2
ENSMUST00000149199.1 ENSMUST00000035056.7 ENSMUST00000148287.1 ENSMUST00000127744.1 |
Dhx30
|
DEAH (Asp-Glu-Ala-His) box polypeptide 30 |
| chr4_-_132422484 | 0.24 |
ENSMUST00000102568.3
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr4_-_129696817 | 0.23 |
ENSMUST00000102588.3
|
Tmem39b
|
transmembrane protein 39b |
| chr7_+_79500081 | 0.23 |
ENSMUST00000181511.2
ENSMUST00000182937.1 |
AI854517
|
expressed sequence AI854517 |
| chr18_+_80046854 | 0.16 |
ENSMUST00000070219.7
|
Pard6g
|
par-6 partitioning defective 6 homolog gamma (C. elegans) |
| chr12_+_49385174 | 0.16 |
ENSMUST00000110746.1
|
3110039M20Rik
|
RIKEN cDNA 3110039M20 gene |
| chr13_-_106936907 | 0.15 |
ENSMUST00000080856.7
|
Ipo11
|
importin 11 |
| chr2_+_85037448 | 0.15 |
ENSMUST00000168266.1
ENSMUST00000130729.1 |
Ssrp1
|
structure specific recognition protein 1 |
| chr3_-_123690806 | 0.15 |
ENSMUST00000154668.1
|
Ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_+_42697146 | 0.13 |
ENSMUST00000054883.2
|
Pou3f3
|
POU domain, class 3, transcription factor 3 |
| chr9_-_109059711 | 0.10 |
ENSMUST00000061973.4
|
Trex1
|
three prime repair exonuclease 1 |
| chr18_-_10030017 | 0.09 |
ENSMUST00000116669.1
ENSMUST00000092096.6 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr9_+_110333402 | 0.08 |
ENSMUST00000133114.1
ENSMUST00000125759.1 |
Scap
|
SREBF chaperone |
| chr4_+_116221689 | 0.07 |
ENSMUST00000106490.2
|
Pik3r3
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 3 (p55) |
| chr2_-_144332146 | 0.04 |
ENSMUST00000037423.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr17_+_34898463 | 0.03 |
ENSMUST00000114033.2
ENSMUST00000078061.6 |
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2 |
| chr4_+_99656299 | 0.03 |
ENSMUST00000087285.3
|
Foxd3
|
forkhead box D3 |
| chr14_+_75845296 | 0.02 |
ENSMUST00000142061.1
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr2_-_157279519 | 0.01 |
ENSMUST00000143663.1
|
Mroh8
|
maestro heat-like repeat family member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nr0b1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.6 | 2.3 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 5.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.4 | 1.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.4 | 2.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 2.0 | GO:0007354 | zygotic determination of anterior/posterior axis, embryo(GO:0007354) mesodermal cell migration(GO:0008078) establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 | 0.7 | GO:1904057 | negative regulation of sensory perception of pain(GO:1904057) |
| 0.2 | 3.2 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.2 | 2.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 | 1.4 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.2 | 0.6 | GO:1990167 | protein K6-linked deubiquitination(GO:0044313) protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 0.6 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.4 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.1 | 1.0 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 1.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.9 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.3 | GO:2000793 | negative regulation of cell proliferation involved in heart morphogenesis(GO:2000137) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.9 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.1 | 2.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.1 | 4.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.0 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 1.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.1 | 0.6 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.6 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 2.8 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 1.3 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 3.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.6 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 1.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.0 | 0.5 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 1.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.9 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0003299 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.1 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.2 | 2.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.7 | GO:0002111 | BRCA2-BRAF35 complex(GO:0002111) |
| 0.2 | 2.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 3.2 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.4 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.4 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 4.0 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.3 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 1.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 2.8 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 1.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.2 | 1.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 5.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.9 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 3.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 0.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 0.9 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 4.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 2.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) BMP receptor activity(GO:0098821) |
| 0.0 | 2.0 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 3.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 3.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.2 | 3.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 2.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 2.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 3.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |